Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(2):186-198. doi:10.7150/ijbs.10634 This issue Cite
Research Paper
FOXM1 Promotes Lung Adenocarcinoma Invasion and Metastasis by Upregulating SNAIL
1. Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai 200032, China
2. Institute of Pathology, Fudan University, Shanghai 200032, China
3. Cancer Institute, Fudan University Shanghai Cancer Center, Shanghai, 200032, China
4. Institute of Biomedical Sciences, Fudan University, Shanghai 200032, China.
5. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, China
6. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China
7. Department of neurosurgery, 1st affiliated hospital of Sun Yat-sen University, Guangzhou, 510080, China
* Ping Wei, Nu Zhang and Yiqin Wang contributed equally to this work
Received 2014-9-25; Accepted 2014-11-17; Published 2015-1-5
Abstract
The forkhead box M1 (FOXM1) transcription factor is one of the key genes inducing tumor invasion and metastasis by an unknown mechanism. In this study, we set out to investigate the effects of FOXM1 overexpression on metastatic human lung adenocarcinoma and the underlying mechanism. FOXM1 expression was analyzed in 78 frozen lung adenocarcinoma tissue samples using an Affymetrix microarray and a 155-paraffin-embedded lung adenocarcinoma tissue microarray with immunohistochemical detection. FOXM1 was found to be overexpressed in lung adenocarcinoma, particularly in metastatic patients, compared to non-metastatic patients. Knockdown of FOXM1 by a specific siRNA significantly suppressed EMT progression, migration and invasion of lung adenocarcinoma cells in vitro, and tumor growth and metastasis in vivo, whereas restored expression of FOXM1 had the opposite effect. FOXM1 binds directly to the SNAIL promoter through two specific binding sites and constitutively transactivates it. Collectively, our findings indicate that FOXM1 may play an important role in advancing lung adenocarcinoma progression. Aberrant FOXM1 expression directly and constitutively activates SNAIL, thereby promoting lung adenocarcinoma metastasis. Inhibition of FOXM1-SNAIL signaling may present an ideal target for future treatment.
Keywords: Lung adenocarcinoma, Invasion, Metastasis, FOXM1, SNAIL
Introduction
Lung cancer is currently the most frequent cause of major cancer incidence and mortality worldwide (1). Two main histological categories are included: small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). NSCLC comprises about 80% of all diagnosed lung cancer, and lung adenocarcinoma is the most common subtype of NSCLC (2). Among patients with lung cancer, 70% present with advanced-stage disease when diagnosed, which has already spread locally or to distant organs (3). While targeted therapy, based on patient-specific molecular aberrations, has recently led to significant responses in lung adenocarcinomas (4, 5), about half of the lung cancer patients do not harbor known driver mutations and cannot be treated with targeted therapy (6). Therefore, novel insights into the molecular mechanisms underlying lung adenocarcinoma pathogenesis as well as regulators and potential targets for treatment are critically needed.
FOXM1, a transcriptional factor of the forkhead box family, is involved in the regulation of organism development and cell proliferation (7, 8). FOXM1 is considered to be a key regulator of both G1-S and G2-M phases of the cell cycle and mitotic spindle integrity (9). Furthermore, FOXM1 is aberrantly overexpressed in the majority of human solid cancers and highly associated with poor clinical prognosis, including glioma, colon cancer, and hepatocellular carcinomas (10-13). Recent studies indicated that aberrant FoxM1 drives the overexpression of some oncogenes and plays an important role in angiogenesis, cell cycle acceleration, metastasis, and drug resistance (14-16). We have previously reported that FOXM1 activates Wnt signaling in glioma stem cells and induces self-renewal (17). In addition, abnormal expression of FOXM1 leads to overexpression of plasminogen activator, urokinase receptor (PLAUR), and this novel FOXM1-PLAUR signaling plays an important role in colon cancer progression and metastasis (18). FOXM1 transcriptionally regulates the expression of the Cav-1 gene and positively transactivates it, promoting pancreatic cancer epithelial-mesenchymal transition (EMT), invasion, and metastasis (19). FOXM1 promotes breast cancer metastasis by increasing nuclear retention of SMAD3 (20). Interestingly, FOXM1 is aberrantly expressed in non-small cell lung carcinomas, suggesting it plays an important role in lung carcinogenesis (21). However, the precise function of FOXM1 and the underlying mechanisms of its activity in lung adenocarcinoma EMT and metastasis remain unclear.
SNAIL is the key regulator of EMT, inducing it by partially repressing epithelial markers and up-regulating mesenchymal markers (22, 23). SNAIL is a member of the Snail family of zinc finger transcription factors (24), aberrantly overexpressed in multiple malignant tumors, including colon cancer, hepatocellular carcinoma, and breast cancer (25-27). The expression of SNAIL is positively associated with a more aggressive phenotype, higher risk for distant metastases and poorer survival rates (28, 29). Furthermore, SNAIL could promote an invasive phenotype in lung carcinoma, whereas the underlying mechanisms are unknown (30). In the present study, we sought to determine the role of FOXM1 in lung adenocarcinoma invasion and metastasis and its molecular mechanism. We identified that FOXM1 directly regulates SNAIL expression in lung adenocarcinoma and sequentially promotes tumor metastasis and invasion. This important FOXM1-SNAIL signaling may be used for future target therapy of lung adenocarcinoma.
Materials and Methods
Human sample specimens and immunohistochemistry analysis
The use of human lung adenocarcinoma specimens was approved by the institutional review board of The Shanghai Cancer Center at Fudan University. 78 frozen lung adenocarcinoma tissue samples were used in the gene chip microarray assay, including 21 grade I, 21 grade II and 36 grade III. In the grade I group, 7 patients are metastatic. In the grade II group, 12 patients are metastatic. In the grade III group, 26 patients are metastatic, respectively. 155-paraffin-embedded lung adenocarcinoma tissue samples were used in the tissue microarray assay, including 51 grade I, 41 grade II and 63 grade III. In the grade I group, 15 patients are metastatic. In the grade II group, 23 patients are metastatic. In the grade III group, 46 patients are metastatic, respectively. We stained tissue sections (4 µm) from paraffin-embedded lung adenocarcinoma specimens with an antibody against human FOXM1 (Santa Cruz Biotechnology, Dallas, TX, USA) or an anti-human SNAIL antibody (Abgent, San Diego, CA, USA). For negative controls, the primary antibody was replaced with control IgG (Santa Cruz Biotechnology, Dallas, TX, USA). Staining results were scored by two investigators blinded to the clinical data. We quantitatively scored the tissue sections according to percentage of positive cells and staining intensity, as previously described (31).
Cell lines and culture conditions
The human lung cancer cell lines NCI-H1650, H1299, A549, HCC827, NCI-H358, SPCA1, and H460 were obtained from the China Center for Type Culture Collection. Lung cancer cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal bovine serum (FBS), sodium pyruvate, nonessential amino acids, and L-glutamine (Life Technologies, Carlsbad, CA, USA).
Transient or stable transfection of lung adenocarcinoma cell lines
To overexpress FOXM1, we transfected NCI-H358 and HCC827 cells with pcDNA3.1-FOXM1 or control vector pcDNA3.1 plasmids (32). Stably transfected cell lines were isolated by selection with G418 (500 µg/mL; Life Technologies, Carlsbad, CA, USA). To inhibit FOXM1 expression, we transfected A549 and NCI-H1650 cells with a FOXM1 siRNA oligonucleotide of the sequence CUCUUCUCCCUCAGAUAUAdTdT (17) or with control siRNA. Sh-FOXM1 lentiviruses were generated with oligonucleotides (5′-ggatcccgctcttctccctcagatatattgatatccgtatatctgagggagaagagttttttccaactcgag-3′). To establish stable cell lines, Sh-FOXM1 lentiviruses was transduced into lung adenocarcinoma cells with polybrene (6 µg/mL; Sigma-Aldrich, St. Louis, MO, USA). Cells were then selected with 10 µg/mL puromycin for 14 days.
Western blot assay
Standard Western blotting was carried out using whole-cell protein lysates of lung adenocarcinoma cells and primary antibodies against FOXM1 (K-19, Santa Cruz Biotechnology, USA), E-cadherin (EPR699, Abcam, Cambridge, UK), N-cadherin(EPR1791-4, Abcam, Cambridge, UK), vimentin (EPR3776, Abcam, Cambridge, UK), and SNAIL (RB1400, Abgent, USA) and secondary antibodies (anti-rabbit IgG or anti-mouse IgG; Cell Signaling Technology, Danvers, MA, USA). Equal-amount protein sample loading was monitored using an anti-Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (EPR6256, Abcam, Cambridge, UK).
Reverse transcription (RT) and real-time PCR
Total RNA was isolated from cells using TRIzol reagent (Life Technologies, USA). First-strand cDNA was synthesized from 1 ug of total RNA using Moloney murine leukemia virus reverse transcriptase (Life Technologies, USA). Real-time PCR analysis of gene expression was carried out using 2 uL of cDNA and the SYBR Green Master Mix (Promega, Madison, WI, USA), as recommended by the manufacturer. GAPDH was used as an internal control. PCR products were run in triplicate for target and internal control genes.
FOXM1 primers 5′-AAGGTTGAGGAGCCTTCGAG-3′ (sense) and 5′-ATTCGGTCGTTTCTGCTGCTT-3′ (antisense); SNAIL primers 5′-CCTCCCTGTCAGATGAGGAC-3′ (sense) and 5′-CCAGGCTGAGGTATTCCTTG-3′ (antisense); E-cadherin primers 5′-TGCCCAGAAAATGAAAAAGG-3′ (sense) and 5′-GTGTATGTGGCAATGCGTTC-3′ (antisense); Vimentin primers 5′-GAGAACTTTGCCGTTGAAGC-3′ (sense) and 5′-GCTTCCTGTAGGTGGCAATC-3′ (antisense); N-cadherin primers 5′-ACAGTGGCCACCTACAAAGG-3′ (sense) and 5′-CCGAGATGGGGTTGATAATG-3′ (antisense); GAPDH primers 5′-CGGATTTGGTCGTATTGG-3′ (sense) and 5′-TCCTG GAAGATGGTGATG-3′ (antisense);
Cell scratch-wound assay
Lung adenocarcinoma cells were grown in 6-well plates until confluence. A cell scratch-wound was generated by scraping with a 10-µL tip. After 12 h, the wounded cells were photographed, and cell migration was assessed by measuring gap sizes in multiple fields.
Cell migration and invasion assays
Cell migration assays were conducted using a polycarbonate membrane pre-coated with Matrigel (24-well Transwell; Corning, Corning, NY, USA), and cell invasion assays were performed using a modified 24-well Boyden chamber with a membrane that was pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA), respectively. Briefly, 24 hours after transfection of NCI-H1650 cells with FOXM1 siRNA and NCI-H358 cells with pcDNA3.1-FOXM1, or their respective controls, 500 µL of cells (1 × 105/mL) in serum-free medium were loaded into the upper wells, while medium containing 20% FBS was placed in the lower wells as a chemoattractant. Cells on the bottom surface of filters were fixed, stained with gentian violet, and counted microscopically in five randomly selected fields at 200× magnification.
Cell immunofluorescence
A549, NCI-H1650, HCC827, and NCI-H358 cells were cultured on Falcon chamber slides (BD Biosciences, USA) until 50-60% confluence before being fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. Cells were immersed three times with cold phosphate-buffered saline (PBS), incubated with FOXM1 (Santa Cruz Biotechnology, USA), E-cadherin, N-cadherin, vimentin (Abcam, Cambridge, UK), and SNAIL (Abgent, USA) primary antibodies at 4°C overnight, subsequently incubated with corresponding Alexa Fluor-conjugated secondary antibodies (Life Technologies, USA) at room temperature for 1 h, and mounted using ProLong® Gold Antifade Reagent with DAPI (Life Technologies, USA). Microscopic images of cells were obtained using a Leica inverted fluorescence microscope (Leica Microsystems, Wetzlar, Germany) with ProgRes Image Capture Software (JENOPTIK Optical Systems, Jena, Germany) and a Leica Confocal LAS-AF SP5 System (Leica Microsystems, Germany).
Promoter reporter and dual luciferase assay
A 2.0-kb SNAIL promoter was cloned into pGL3-Basic Luciferase Reporter Vectors (Promega, USA). Site-specific mutagenesis of the SNAIL promoter was carried out using a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA), according to the manufacturer's instructions. Primers used to generate the mutant vector were as follows: mut1, 5′-TGTGAGGTTTATGCCAGAGCCACCC-3′ (sense) and 5′-GGGTGGCTCTGGCATAAACCTCACA-3′ (antisense); mut2, 5′-GTAATTATCTGTGCACTTCGTCTGTC-3′ (sense) and 5′-GACAGACGAAGTGCACAGATAATTAC-3′ (antisense); mut1and2, 5′-TGTGAGGTTTATGCCAGAGCCACCC-3′ (sense) and 5′-GACAGACGAAGTGCACAGATAATTAC-3′ (antisense). The mutation was confirmed by DNA sequencing. SNAIL promoter activity was normalized by cotransfection with a ®-actin/Renilla luciferase reporter, containing a full-length Renilla luciferase gene (31). Both, firefly and Renilla luciferase activity were quantified using a Dual-Luciferase Reporter Assay System (Promega, USA) 24 h after transfection.
Chromatin immunoprecipitation (ChIP) assay
Tumor cells (5 × 106) were prepared for the chromatin immunoprecipitation (ChIP) assay with the ChIP assay kit (Millipore, Billerica, MA, USA), according to the manufacturer's protocol. The resulting precipitated DNA samples were analyzed using PCR to amplify a potential binding site 1 region of the SNAIL promoter with the primers 5′-AGACAGTAGTTCTGCCCTTCAGGTT-3′ (sense) and 5′-ATGGAGCCGTGTTACAGCCT-3′ (antisense), and a potential binding site 2 region of the SNAIL promoter with the primers 5′-AGTTGCCACTTCTTCCCTCGGGCCT-3′ (sense) and 5′-GGAACGGGTGCTCTTGGCT-3′ (antisense). PCR products were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining.
Animal experiments
All procedures involving mice were conducted in accordance with Fudan University Shanghai Cancer Center Animal Care guidelines. All efforts were made to minimize animal suffering, to reduce the number of animals used, and to utilize possible alternatives to in vivo techniques. Tumor cells in the exponential growth phase were harvested by brief exposure to 0.25% trypsin/0.02% EDTA solution (w/v). Cell viability was determined using Trypan blue exclusion, and only single-cell suspensions that were more than 95% viable were used. Groups of five nude mice were injected with tumor cells either subcutaneously (1 × 106 per mouse) or into the tail vein (5 × 106 per mouse). Subcutaneously injected animals were killed 6 weeks later or when they had become moribund, and tumors were removed and weighed. Tail-injected animals were killed 4 weeks after the injection or when they had become moribund, their lungs were removed, and surface metastases were counted. Every surface was examined by two investigators who were unaware of the experimental protocol and scored separately. Tissue was fixed in 10% buffered formalin, immersed in an ascending series of alcohols, and paraffin embedded. 4 µm sections were cut and stained with hematoxylin and eosin (H & E).
Statistical analysis
The significance of the data from patient specimens was determined by the Pearson correlation coefficient. The 2-tailed χ2 test was used to determine the significance of differences between covariates. Survival durations were calculated using the Kaplan-Meier method. The log-rank test was used to compare cumulative survival rates in patient groups. The significance of in vitro and in vivo data was determined by Student's t-test (two-tailed), considering P < 0.05 as significant.
Results
FOXM1 expression in human lung adenocarcinoma specimens and its association with lung cancer pathologic features
To screen for novel molecular events that lead to metastasis of lung adenocarcinoma, genome-wide gene expression profiling was conducted on 78 frozen lung adenocarcinoma samples, using the Affymetrix GeneChip® Human Genome U133 Plus 2.0 microarray. FOXM1 expression was elevated in the stage II and III groups (I vs. II, P < 0.05; I vs. III, P < 0.01; Fig. 1, left panel), as well as in the metastasis group (P < 0.05; Fig. 1, right panel). These results were confirmed by semiquantitative RT-PCR experiments using the same 78 lung adenocarcinoma samples (Supplementary Material: Fig. S1).
To further evaluate the effect of FOXM1 expression on lung adenocarcinoma progression and metastasis, we investigated the expression of FOXM1 protein with the 155-primary lung adenocarcinoma tissue microarray from stages I to III. We observed FOXM1-positive staining in the nuclei of tumor cells and FOXM1-negative or weak FOXM1-positive staining in adjacent normal lung cells (Fig. 1, B). Increasing FOXM1 expression correlated with decreasing levels of tumor differentiation, indicating that FOXM1 expression is significantly upregulated in poorly differentiated tumors (Fig. 1, C). In addition, FOXM1 expression was positively correlated with disease stages, particularly in early-stage (stage I) and late-stage (stage III) tumors (Fig. 1, D). Moreover, FOXM1 expression in distant metastasis specimens was dramatically higher than in non-metastasis specimens (Fig. 1, E). Correlations between FOXM1 expression and clinicopathologic characteristics are summarized in Supplementary Material: Table S1, showing that increased FOXM1 expression is correlated with disease stage (P = 0.001), pT classification (P = 0.001), distant metastasis (P = 0.001), vessel invasion (P = 0.002), and disease differentiation (P = 0.001). These findings indicate that FOXM1 expression plays a critical role in lung adenocarcinoma development and progression and is a valuable biomarker for this disease.
FOXM1 expression in lung adenocarcinoma specimens and its association with lung adenocarcinoma pathologic features. A, Genome-wide gene expression profiling was conducted on 78 frozen lung adenocarcinoma samples, using the Affymetrix GeneChip® Human Genome U133 Plus 2.0 microarray. FOXM1 expression was elevated in the stage II and III groups (I vs. II, P < 0.05; I vs. III, P < 0.01; left panel), as well as in the metastasis group (P < 0.05; right panel). B, Tissue microarray immunostaining using a specific antibody against FOXM1. Representative images of FOXM1 protein expression in adjacent normal lung tissue, lung adenocarcinoma tissue, and lymph node tissue are shown (200×). Note that the majority of adjacent normal lung tissue cells are negative for FOXM1 expression, whereas lung adenocarcinoma tissue cells and lymph node cells are strongly positive for FOXM1 expression. C, FOXM1 expression is positively correlated with tumor differentiation, and representative images of high and low grade tumors are shown (200 x). The numbers of samples of low and high grade were 67 and 88, respectively. D, FOXM1 expression was positively correlated with disease stage, and representative images of stages I and III are shown (200×). The sample numbers of stages I, II, and III were 51, 41, and 63, respectively. E, FOXM1 expression was positively correlated with tumor metastasis, and representative images of tumors from patients with or without distant metastases are shown (200×). The numbers of patients with or without distant metastases were 84 or 71, respectively. F, DFS (left) and OS (right) rates were estimated using the Kaplan-Meier method. Both rates were significantly worse in patients with FOXM1-positive primary tumors than in patients with FOXM1-negative primary tumors (P < 0.001 for DFS, P = 0.003 for OS).
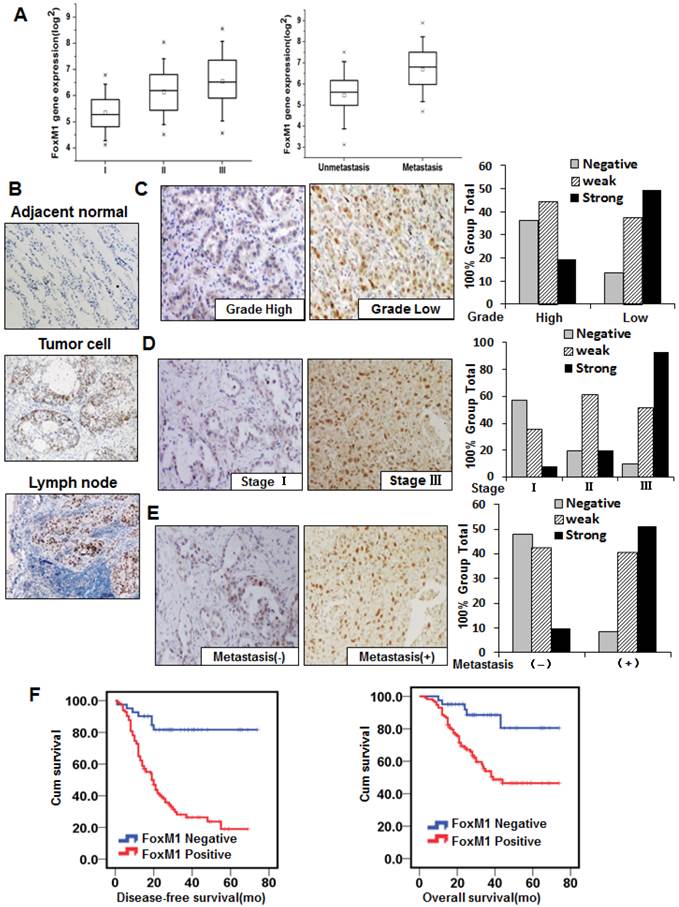
To assess the clinical significance of FOXM1 overexpression in lung adenocarcinoma, we scored FOXM1 staining with the 155-primary lung adenocarcinoma tissue microarray as previously described (31). There was a significant difference between the FOXM1-positive and FOXM1-negative groups in the number of patients who developed metastasis from primary lung adenocarcinoma after surgery. Positive FOXM1 expression was associated with a significantly increased risk of distant metastases (HR 5.739; 95% CI, 2.64-12.47; P < 0.001; Fig. 1, left panel). Moreover, the Overall Survival (OS) was significantly higher in patients with FOXM1-negative tumors than in those with FOXM1-positive tumors (HR 4.06; 95% CI, 1.62-10.20; P = 0.003; Fig. 1, right panel), indicating that FOXM1 is a negative total survival marker for lung adenocarcinoma patients. These results strongly indicate that FOXM1 expression is directly associated with metastasis and prognosis of patients with lung adenocarcinoma.
Altered FOXM1 expression affects EMT induction in human lung adenocarcinoma cells
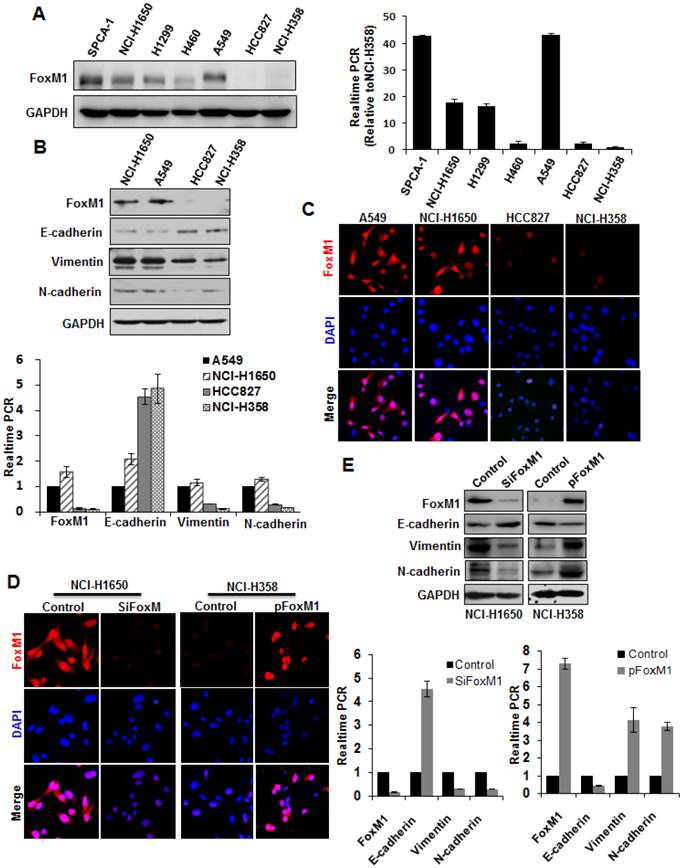
As the overexpression of FOXM1, examined by immunohistochemistry, was associated with lung adenocarcinoma metastasis and ascending clinical stage, we subsequently investigated whether FOXM1 was capable of inducing EMT in human lung adenocarcinoma cells. First, we screened expression levels of FOXM1 in different lung adenocarcinoma cell lines (Fig. 2). Then, we chose two cell lines expressing high levels of FOXM1 (A549, NCI-H1650) and two expressing low levels (HCC827, NCI-H358), and carried out Western blot (Fig. 2, upper panel), real-time PCR (Fig. 2, lower panel) and cell immunofluorescence (Fig. 2, C) analyses with these four cell lines. We found that FOXM1 expression correlated with the expression levels of EMT markers: the expression of an epithelial marker (E-cadherin) was higher in the low-level FOXM1-expressing cells (HCC827, NCI-H358) than in the high-level FOXM1-expressing cells (A549, NCI-H1650; Fig. 2). In contrast, the expression of mesenchymal markers (vimentin, N-cadherin) was lower in the low-level FOXM1-expressing cells (HCC827, NCI-H358) than in the high-level FOXM1-expressing cells (A549, NCI-H1650; Fig. 2).
To determine the effect of altered FOXM1 expression on EMT induction in lung adenocarcinoma cells, we transfected the FOXM1 expression vector pcDNA3.1-FOXM1 (or the control vector) into NCI-H358 cells. We found that elevated expression of FOXM1 in NCI-H358 cells (Fig. 2, D) significantly increased vimentin and N-cadherin expression but decreased E-cadherin expression, both on the mRNA (Fig. 2, lower panel) and protein level (Fig. 2, upper panel). In contrast, FOXM1-specific siRNA led to a concomitant decrease in vimentin and N-cadherin but increase in E-cadherin expression, both on the mRNA (Fig. 2, D; Fig. 2, E lower panel) and protein level (Fig. 2, E upper panel) in NCI-H1650 cells.
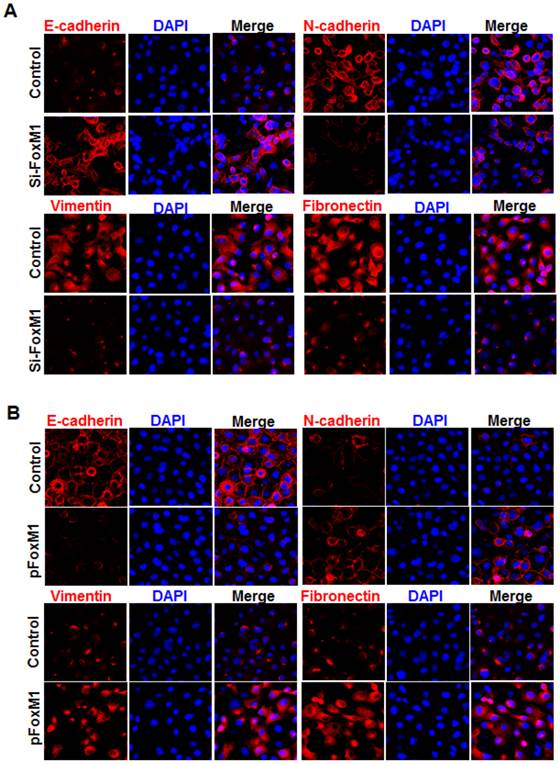
Immunofluorescence staining further revealed that E-cadherin (Fig. 3, A) was strongly downregulated in FOXM1 siRNA-transfected NCI-H1650 cells, while levels of N-cadherin, vimentin and fibronectin were dramatically induced in FOXM1 silenced NCI-H1650 cells (Fig. 3, A). Ectopic expression of FOXM1 in NCI-H358 cells also decreased the levels of E-cadherin (Fig. 3, B) and increased the levels of N-cadherin, vimentin and fibronectin (Fig. 3, B). These results indicate that altered FOXM1 expression affects EMT induction in lung adenocarcinoma cells.
Altered FOXM1 expression affects migration and invasion of human lung adenocarcinoma cells in vitro
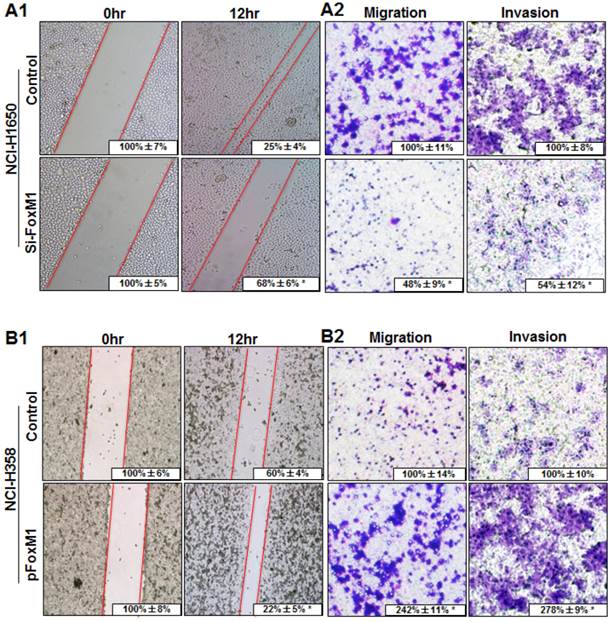
To determine the effect of altered FOXM1 expression on migration and invasion of lung adenocarcinoma cells, FOXM1-transfected and FOXM1-siRNA-transfected lung adenocarcinoma cells were wounded by scratching and maintained at 37°C for additional 12 h. Down-regulation of FOXM1 attenuated the flattening and spreading of NCI-H1650 cells, whereas ectopic FOXM1 expression strongly promoted the flattening and spreading of NCI-H358 cells (Fig. 4, A1, B1). These results were confirmed by migration and invasion assays, revealing that the migratory and invasive ability were attenuated in FOXM1-siRNA-transfected NCI-H1650 cells, whereas the levels of both migration and invasion of FOXM1-transfected NCI-H358 cells were significantly higher than those of control cells (Fig. 4, A2, B2).
Effects of altered FOXM1 expression on tumorigenicity and metastasis of human lung adenocarcinoma cells in vivo
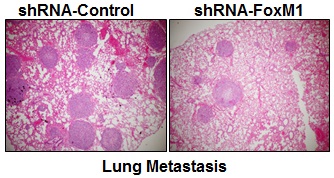
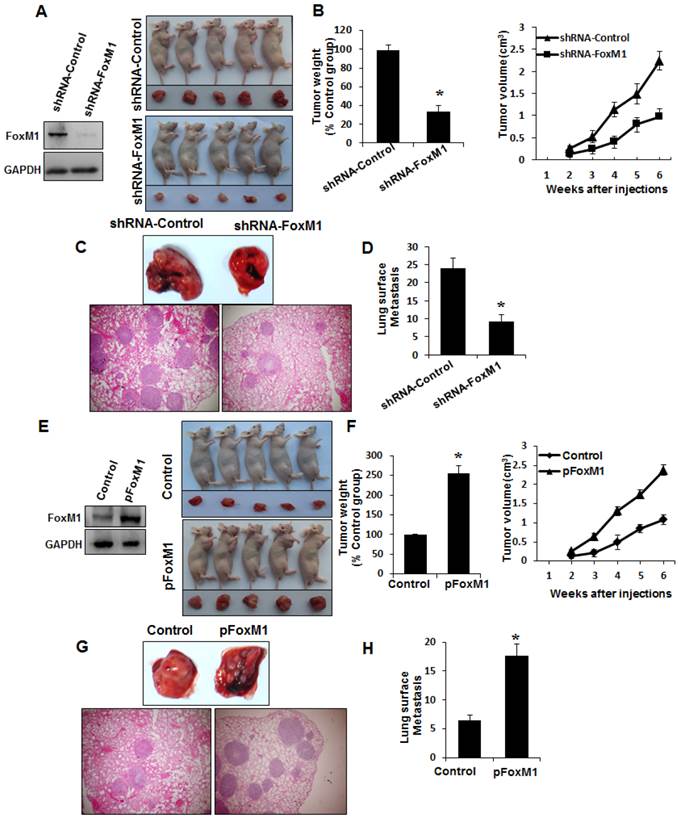
To determine whether FOXM1 plays an important role in the tumorigenicity and metastasis of human lung adenocarcinoma cells in vivo, we injected FOXM1-shRNA-transfected A549 cells into either the subcutis or the caudal vein of nude mice in groups of five. Consistent with the effect of altered FOXM1 expression on migration and invasion of colon cancer cells in vitro, knockdown of FOXM1 significantly inhibited lung tumor growth (Fig. 5, A, B) and abrogated lung metastases (Fig. 5, C, D) of A549 cells in nude mice, whereas overexpression of FOXM1 significantly promoted tumor growth (Fig. 5, E, F) and increased lung metastases (Fig. 5, G, H) of NCI-H358 cells. Our data clearly indicate that FOXM1 is oncogenic and promotes tumorigenicity and metastasis of lung adenocarcinoma in vivo.
Correlation between FOXM1 expression and EMT potential of lung adenocarcinoma cells. A, Western blot (left panel) and real-time PCR (right lanel) analysis of FOXM1 protein expression in lung adenocarcinoma cell lines. Quantitative real-time PCR results were standardized in relation to GAPDH and expressed as multiples of NCI-H358 results. B, Western blot (upper panel) and real-time PCR (lower panel) analysis of expression levels of FOXM1, E-cadherin, vimentin, and N-cadherin. Note that high-level FOXM1-expressing cells (NCI-H1650 and A549) showed high-level expression of mesenchymal markers (vimentin, N-cadherin) and low-level expression of epithelial markers (E-cadherin), whereas low-level FOXM1-expressing cells (HCC827 and NCI-H358) showed the opposite. C, Cell immunofluorescence staining for FOXM1 (red) and nuclei (DAPI; blue). Note that NCI-H1650 and A549 cells displayed strong positive FOXM1 staining, whereas HCC827 and NCI-H358 cells displayed weak positive FOXM1 staining. D, Cell immunofluorescence staining for FOXM1 (red) and nuclei (DAPI; blue). NCI-H1650 cells were transfected with control siRNA (“control”) or siRNA against FOXM1 (“siFOXM1”), whereas NCI-H358 cells were transfected with control pcDNA3.1 vector (“control”) or pcDNA3.1-FOXM1 expression vector (“p FOXM1”). E, Western blots (upper panel) and real-time PCR (lower panel) analysis of EMT markers. Total protein lysates or RNA were prepared from NCI-H1650 cells with FOXM1 knockdown or NCI-H358 cells with FOXM1 overexpression (as described in D).
A close relationship between altered expression of FOXM1 and SNAIL in lung adenocarcinoma
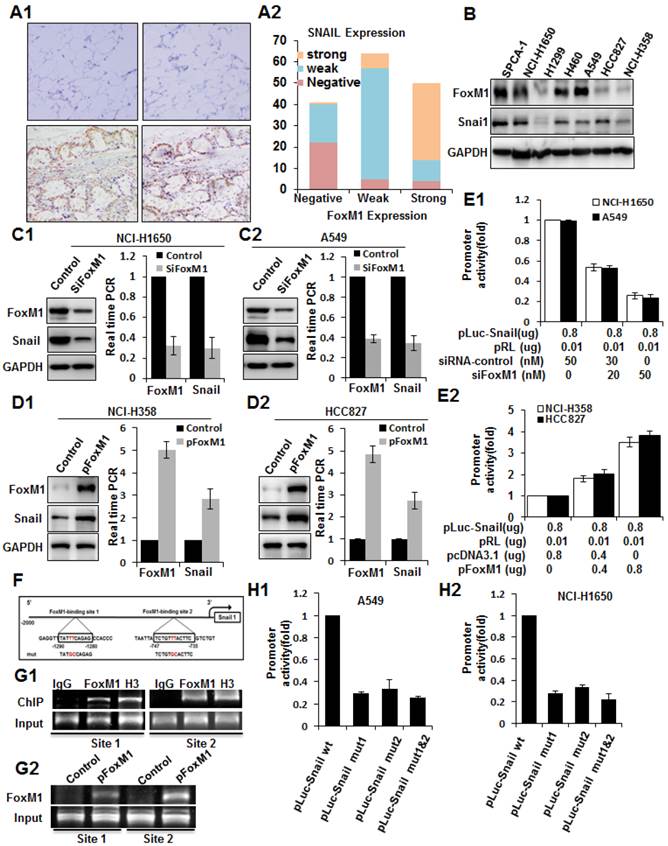
To explore the mechanisms underlying FOXM1 overexpression in lung adenocarcinoma, we analyzed both FOXM1 and SNAIL expression in tissues and cell lines. By analyzing consecutive primary lung adenocarcinoma sections, we observed that FOXM1 expression was significantly correlated with SNAIL expression. Lung adenocarcinoma, but not adjacent tissues, expressed both FOXM1 and SNAIL (Fig. 6, A1), and their direct correlation was found statistically significant (r = 0.645; P < 0.01; Fig. 6, A2). Consistently, the expression of FOXM1 also correlated with the expression of SNAIL in lung adenocarcinoma cell lines (Fig. 6, B).
To further explore the molecular mechanisms of regulation of SNAIL expression by FOXM1, we first determined the impact of altered FOXM1 expression on SNAIL expression in human lung adenocarcinoma cell lines. We found that knockdown of FOXM1 expression in NCI-H1650 and A549 cells led to significantly decreased SNAIL mRNA and protein expression (Fig. 6, C1, C2). Conversely, increased expression of FOXM1 in NCI-H358 and HCC827 cells led to significantly increased SNAIL mRNA and protein levels (Fig. 6, D1, D2). Furthermore, to determine the role of FOXM1 in regulating SNAIL transcription, we generated the SNAIL promoter pLuc-SNAIL and cotransfected it together with FOXM1 siRNA into NCI-H1650 and A549 cells, causing knockdown of FOXM1 and suppression of the SNAIL promoter in both cell lines (Fig. 6, E1). Conversely, overexpression of FOXM1 after cotransfection of pcDNA3.1-FOXM1 together with pLuc-SNAIL into NCI-H358 and HCC827 cells activated the SNAIL promoter (Fig. 6, E2). Collectively, our findings indicate that FOXM1 regulates SNAIL expression, most likely at the transcriptional level.
Influence of FOXM1 expression on EMT marker alteration of lung adenocarcinoma cells. Cell immunofluorescence staining for E-cadherin (red), N-cadherin (red), vimentin (red), fibronectin (red), and nuclei (DAPI; blue), respectively, using NCI-H1650 cells with FOXM1 knockdown (A) or NCI-H358 cells with FOXM1 overexpression (B).
Influence of FOXM1 expression on lung adenocarcinoma cell migration and invasion. NCI-H1650 (A) and NCI-H358 cells (B) were transfected with FoxM1 siRNA and pcDNA3.1-FoxM1 for 48 h, respectively. For the cell scratch-wound assay, NCI-H1650 (A1) and NCI-H358 cells (B1) were wounded by scratching and maintained at 37°C for additional 12 h. Cells were photographed, and cell migration was assessed by measuring gap sizes (inserted numbers representing percentage area of gap ± SD). Migration and invasiveness of NCI-H1650 (A2) and NCI-H358 cells (B2) cells were determined as described in Materials and Methods. Representative migrating or invading tumor cells were photographed. Data represent the mean ± SD of triplicates (P < 0.01 in a comparison of the FoxM1 siRNA-treated group or the pcDNA3.1-FoxM1-treated group with control groups).
Direct transcriptional regulation of SNAIL expression by FOXM1 in lung adenocarcinoma cells
To determine whether SNAIL could be a direct transcriptional target of FOXM1, we scanned approximately 2-kb of the promoter region of SNAIL with the FOXM1 DNA-binding consensus sequence (31) and found two FOXM1 putative binding sites (-747 to -735 and -1290 to -1280) (Fig. 6, F). To confirm that FOXM1 binds directly to the human SNAIL promoter, we performed ChIP assays using two pairs of PCR primers for sequences in the vicinity of these potential FOXM1 binding sites. Indeed, ChIP assays in NCI-H1650 cells, using antibodies specific to FOXM1, showed that FOXM1 protein binds directly to the endogenous SNAIL promoter in both sites (Fig. 6, G1). Moreover, in NCI-H358 cells, normally expressing low FOXM1 levels, there is an increased binding of FOXM1 protein to the SNAIL promoter after transfection with pcDNA3.1-FOXM1 (Fig. 6, G2). These results were further confirmed by promoter mutation assays. Various mutant reporters were generated from the wild-type SNAIL promoter construct, including a FOXM1-binding site 1 mutation only (pGL3-SNAIL Mut1), a FOXM1-binding site 2 mutation only (pGL3-SNAIL Mut2), and a mutation of both site 1 and site 2 (pGL3-SNAIL Mut3). We transfected these mutant luciferase reporters into NCI-H1650 or A549 cells and compared their activity with that of the wild-type SNAIL promoter (Fig. 6, H1, H2). Disruption of one or both of the FOXM1-binding sites significantly attenuated SNAIL promoter activity in NCI-H1650 or A549 cells. In summary, these findings show that FOXM1 binds specifically to FOXM1-binding sites in the SNAIL promoter, suggesting that SNAIL is a direct transcriptional target of FOXM1.
Discussion
In this study, we determined the critical role of FOXM1 and SNAIL in lung adenocarcinoma pathogenesis and its underlying mechanisms. We observed that elevated FOXM1 expression was directly correlated with lung adenocarcinoma progression and metastasis. FOXM1-positive staining of lung adenocarcinoma cells could be used to identify a greatly increased risk of metastasis in patients after surgery, which might serve as a valuable prognostic marker. We also found that FOXM1 transcriptionally activates the SNAIL gene, thereby constituting an important signaling pathway that directly affects EMT, invasion, and metastasis of lung adenocarcinoma cells in vitro and in vivo. Collectively, our clinical and mechanistic evidence strongly suggests that dysregulated FOXM1 expression causes abnormal SNAIL expression and critically contributes to lung adenocarcinoma pathogenesis and aggressive behavior.
Influence of FOXM1 expression on lung adenocarcinoma growth and metastasis. NCI-H1650 cells with FoxM1 knockdown (A, B) or NCI-H358 cells with FoxM1 overexpression (C, D) were injected into nude mice (n = 5) either subcutaneously into the right scapular region (1 × 106 per mouse) or into the caudal vein (5 × 106 per mouse). Tumor-bearing mice were sacrificed when they became moribund or after 6 weeks (subcutaneous injection) or after 4 weeks (tail injection). Shown are IB blot of the shFoxM1 knockdown or pFoxM1 overexpress (A, E, left panel), gross tumors (A, E, right panel), gross lung metastatic tumors (C, G, upper panel) and H & E-stained sections of liver (C, G, lower panel); tumor weights (B, F, left panel), tumor volumes (B, F, right panel) and the numbers of lung surface metastases (D and H). P < 0.01 in a comparison of the FoxM1 shRNA-treated group or the pcDNA3.1-FoxM1-treated group with control groups.
Coexpression of FOXM1 and SNAIL in lung adenocarcinoma and direct binding of FOXM1 to the SNAIL promoter. A, Immunohistochemical staining (A1) of lung adenocarcinoma specimens using specific anti-FOXM1 and anti-SNAIL antibodies. Shown are representative images of negative (upper 2 panels) and positive (lower 2 panels) FOXM1 and SNAIL staining of lung adenocarcinoma sections (200 x). FOXM1 expression levels were directly correlated with SNAIL expression levels (r = 0.645; P < 0.01, A2) in primary lung adenocarcinoma, as analyzed using the Pearson test. B, Western blot analysis of FOXM1 and SNAIL protein expression in lung adenocarcinoma cell lines. C, NCI-H1650 (C1) and A549 (C2) cells transfected with FOXM1-siRNA or control siRNA. Western blot (left) and RT-PCR (right) analysis of FOXM1 and SNAIL expression. D, NCI-H358 (D1) and HCC827 (D2) cells transfected with pcDNA3.1-FOXM1 or control pcDNA3.1. Western blot (left) and RT-PCR (right) analysis of FOXM1 and SNAIL expression. E, Effects of FOXM1 expression on SNAIL promoter activity. NCI-H1650 and A549 cells were cotransfected with 0.8 µg of the SNAIL promoter-luciferase construct pLuc-Snail and 0-50 nmol/L FOXM1-siRNA or control siRNA (E1), whereas NCI-H358 and HCC827 cells were cotransfected with 0.8 mg pLuc-Cav and 0-0.8 µg of pcDNA3.1-FOXM1 or pcDNA3.1 (E2). Promoter activities were determined using the Dual-Luciferase Assay Kit. F, Sequences and positions of putative FOXM1-binding elements in the SNAIL promoter. G, Binding of FOXM1 to the SNAIL promoter in vivo. ChIP assays were done with NCI-H1650 (G1) and NCI-H358 (G2) cells treated with pcDNA3.1-FOXM1 or pcDNA3.1. Chromatin fragments of cells (1% of total cell lysates as input) were immunoprecipitated with an anti-FOXM1 antibody or control IgG and subjected to PCR. H, SNAIL promoter activity with or without mutations in the FOXM1 binding sites, as determined by luciferase activity in A549 (H1) and NCI-H1650 (H2) cells.
Previous evidence had indicated that FOXM1 might serve as a novel prognostic biomarker involved in the tumorigenesis and metastasis of several malignancies (33). Some studies had suggested a potential role of FOXM1 in NSCLC patients, e.g., Yang et al (34) demonstrated that overexpression of FOXM1 was related to poor survival in 69 squamous cell carcinoma patients, not including lung adenocarcinoma. Xu et al (35) found that FOXM1 overexpression was significantly associated with several clinicopathological parameters such as TNM staging, tumor stage, lymph node metastasis and poorer prognosis in 175 NSCLC patients. The results of these studies were not exactly equivalent to ours, maybe due to different study populations, varied sample quality or cell type (NSCLC vs. lung adenocarcinoma). In our current study, FOXM1 expression was first investigated in 78 frozen lung adenocarcinoma tissue samples using Affymetrix microarrays, then confirmed in the same 78 frozen lung adenocarcinoma tissue samples using real-time PCR and with a 155-paraffin-embedded lung adenocarcinoma tissue microarray using immunohistochemistry. We found that elevated expression of FOXM1 was significantly associated with poorer tumor differentiation and advanced disease stage, particularly with distant metastasis and metastasis-free survival time. Thus, we present very important clinical evidence, indicating that FOXM1 promotes lung adenocarcinoma metastasis.
Both our in vivo and in vitro assays supported this notion. Altered FOXM1 expression affected the ability of migration and invasion of lung adenocarcinoma cell lines. Enhanced expression of FOXM1 in lung adenocarcinoma cell lines was associated with much higher tumor growth and metastatic ability, compared to control cells in the mouse model. Our findings are consistent with recent studies, which have indicated a critical role for FOXM1 in tumorigenesis and metastasis of pancreatic, breast and lung cancer cells (19, 20, 36, 37).
Cancer metastasis is a complex process, and its cellular and molecular mechanisms remain to be determined (38, 39). EMT is a critical process providing epithelial-derived tumor cells with the ability of increased migration and invasiveness, contributing to tumor metastasis (40, 41). In the current study, we found that the expression of FOXM1 is much higher in cell lines of mesenchymal phenotype, such as A549, than in cell lines of epithelial phenotype, such as NCI-H358. Elevated FOXM1 expression could contribute to the acquisition of the EMT phenotype by up-regulating mesenchymal cell markers vimentin and N-cadherin and down-regulating the epithelial cell marker E-cadherin, whereas decreased FOXM1 expression could cause the opposite. Our new findings were consistent with recent studies, which have shown a correlation between FOXM1 up-regulation and cancer cell EMT in human pancreatic cancer cells (19) and another correlation between EMT during radiation-induced pulmonary fibrosis and FOXM1 overexpression (42).
Assuming a crucial role of FOXM1 in the metastasis of malignancies, it is necessary to explore the underlying mechanisms responsible for FOXM1 overexpression in lung adenocarcinoma. Previous studies showed that FOXM1 increases nuclear localization of beta-catenin in glioma stem cells (17) and activates TGF-beta signaling in breast cancer cells (20). In lung adenocarcinoma cell lines, these two important pathways may also be induced by FOXM1, as has been implied in other previous studies (43, 44). The results of our current study indicate that SNAIL, which plays an important role in inducing and promoting EMT in epithelial cells (45), is a novel downstream target of FOXM1. We show that the expression levels of FOXM1 correspond directly to the expression levels of SNAIL, both in human lung adenocarcinoma tissues and lung adenocarcinoma cell lines. By using a well-established FOXM1 overexpression and knockdown system, we found that overexpression of FOXM1 led to increased expression of SNAIL, whereas knockdown of FOXM1 had the opposite effect. Moreover, we noted that FOXM1 could transactivate SNAIL promoter activity, in agreement with recent studies, showing that FOXM1 translationally regulated SNAIL during lung inflammation (42), yet not indicating the exact binding site of FOXM1 in the human SNAIL promoter. Our current study specifically identified two potential FOXM1-binding sites in human SNAIL promoters. Mutations in these sites significantly attenuated FOXM1-mediated transactivation of the human SNAIL promoter, and ChIP assays confirmed the recruitment of FOXM1 to these binding sites in the human SNAIL promoter in vitro.
Conclusions
In summary, this study provides both clinical and molecular evidence supporting the regulatory role of FOXM1 in SNAIL expression and a novel FOXM1-SNAIL signaling pathway in tumorigenesis and metastasis of lung adenocarcinoma. Our study not only indicates a very important molecular mechanism of lung adenocarcinoma progression and metastasis, but it also provides a promising molecular target for designing optional therapeutic modalities to control the progression and metastasis of lung adenocarcinoma.
Supplementary Material
Table S1 and Fig.S1.
Acknowledgements
National Natural Science Foundation of China (81101586, 81201836, 81372646, 81370072), Shanghai Pujiang Program (12PJD015, 12PJ1401800), National Basic Research Program of China (973 Program, Grant No. 2013CB733700), Shanghai Science and Technology Commission of Shanghai Municipality (No.10DJ1400500, No.10DJ1400501, 12410707700), the Fundamental Research Funds for the Central Universities (11ykzd06), the Natural Science Foundation of Guangdong Province for Distinguished Young Scholars (S2013050014535), Pearl River New Star Science and technology program of Guangzhou City (2013J2200022), Science and Technology Program of Guangzhou (2013J4100060).
Authors' contributions
All authors participated in the interpretation of the studies, analysis of the data and review of the manuscript. Ping Wei, Xiaoyan Zhou, and Xiang Du participated in the study design and supervision. Ping Wei, Yiqing Wang, and Nu Zhang conducted the experiments. Dawei Li and Lisha Wang conducted in vitro experiments. Xiangjie Sun, Chen Shen and Yusi Yang collected the clinical and follow-up data. Ping Wei wrote the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
1. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90
2. Sharma SV, Bell DW, Settleman J. et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169-81
3. Sangodkar J, Katz S, Melville H. et al. Lung adenocarcinoma: lessons in translation from bench to bedside. Mt Sinai J Med. 2010;77(6):597-605
4. Soda M, Choi YL, Enomoto M. et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561-6
5. Keedy VL, Temin S, Somerfield MR. et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29(15):2121-7
6. Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18(3):349-51
7. Ye H, Kelly TF, Samadani U. et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626-41
8. Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25(9):1715-9
9. Wang IC, Chen YJ, Hughes D. et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875-94
10. Madureira PA, Varshochi R, Constantinidou D. et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281(35):25167-76
11. Kalinichenko VV, Major ML, Wang X. et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18(7):830-50
12. Zhang Y, Zhang N, Dai B. et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68(21):8733-42
13. Li D, Peng Z, Tang H. et al. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011;17(11):3558-68
14. Li Q, Zhang N, Jia Z. et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69(8):3501-9
15. Monteiro LJ, Khongkow P, Kongsema M. et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2013;32(39):4634-45
16. Schimmel J, Eifler K, Sigurethsson JO. et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53(6):1053-66
17. Zhang N, Wei P, Gong A. et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20(4):427-42
18. Li D, Wei P, Peng Z. et al. The critical role of dysregulated FOXM1-PLAUR signaling in human colon cancer progression and metastasis. Clin Cancer Res. 2013;19(1):62-72
19. Huang C, Qiu Z, Wang L. et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72(3):655-65
20. Xue J, Lin X, Chiu WT. et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J Clin Invest. 2014;124(2):564-79
21. Balli D, Zhang Y, Snyder J. et al. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res. 2011;71(1):40-50
22. Batlle E, Sancho E, Franci C. et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84-9
23. Guaita S, Puig I, Franci C. et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277(42):39209-16
24. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3(3):155-66
25. Miyoshi A, Kitajima Y, Kido S. et al. Snail accelerates cancer invasion by upregulating MMP expression and is associated with poor prognosis of hepatocellular carcinoma. Br J Cancer. 2005;92(2):252-8
26. Roy HK, Smyrk TC, Koetsier J. et al. The transcriptional repressor SNAIL is overexpressed in human colon cancer. Dig Dis Sci. 2005;50(1):42-6
27. Come C, Magnino F, Bibeau F. et al. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12(18):5395-402
28. Elloul S, Elstrand MB, Nesland JM. et al. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103(8):1631-43
29. Moody SE, Perez D, Pan TC. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197-209
30. Merikallio H, Turpeenniemi-Hujanen T, Paakko P. et al. Snail promotes an invasive phenotype in lung carcinoma. Respir Res. 2012;13:104
31. Dai B, Kang SH, Gong W. et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26(42):6212-9
32. Liu M, Dai B, Kang SH. et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66(7):3593-602
33. Park HJ, Gusarova G, Wang Z. et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med. 2011;3(1):21-34
34. Yang DK, Son CH, Lee SK. et al. Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Hum Pathol. 2009;40(4):464-70
35. Xu N, Jia D, Chen W. et al. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One. 2013;8(3):e59412
36. Ke Y, Zhao W, Xiong J. et al. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem Res Int. 2013;2013:506731
37. Kong FF, Qu ZQ, Yuan HH. et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31(6):2660-8
38. Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895-904
39. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559-64
40. Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17(5):548-58
41. Hugo H, Ackland ML, Blick T. et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213(2):374-83
42. Balli D, Ustiyan V, Zhang Y. et al. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. Embo J. 2013;32(2):231-44
43. Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106(1):t356
44. Risolino M, Mandia N, Iavarone F. et al. Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-beta-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014
45. Kudo-Saito C, Shirako H, Takeuchi T. et al. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195-206
Author contact
![]() Corresponding authors: Xiang Du, No.2 building, 270 Dong'an Road, Shanghai, 200032. Tel: 86-21-64175590; Fax: 86-21-64175590; Email: dx2008cncom
Corresponding authors: Xiang Du, No.2 building, 270 Dong'an Road, Shanghai, 200032. Tel: 86-21-64175590; Fax: 86-21-64175590; Email: dx2008cncom

 Global reach, higher impact
Global reach, higher impact