Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(6):730-745. doi:10.7150/ijbs.15066 This issue Cite
Research Paper
Different Modulatory Mechanisms of Renal FXYD12 for Na+-K+-ATPase between Two Closely Related Medakas upon Salinity Challenge
1. Department of Life Sciences, National Chung Hsing University, Taichung 402, Taiwan.
2. Tainan Hydraulics Laboratory, National Cheng Kung University, Tainan 709, Taiwan.
3. National Institute for Basic Biology, National Institutes of Natural Sciences, Okazaki, Aichi 444-8787, Japan.
4. Department of Biological Science and Technology, China Medical University, Taichung 404, Taiwan.
Received 2016-1-24; Accepted 2016-3-21; Published 2016-4-28
Abstract
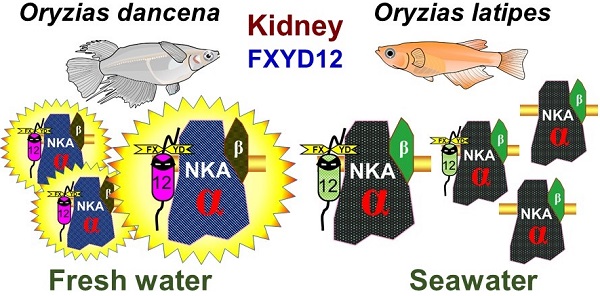
Upon salinity challenge, the Na+-K+-ATPase (NKA) of fish kidney plays a crucial role in maintaining ion and water balance. Moreover, the FXYD protein family was found to be a regulator of NKA. Our preliminary results revealed that fxyd12 was highly expressed in the kidneys of the two closely related euryhaline medaka species (Oryzias dancena and O. latipes) from different natural habitats (brackish water and fresh water). In this study, we investigated the expression and association of renal FXYD12 and NKA α-subunit as well as potential functions of FXYD12 in the two medakas. These findings illustrated and compared the regulatory roles of FXYD12 for NKA in kidneys of the two medakas in response to salinity changes. In this study, at the mRNA and/or protein level, the expression patterns were similar for renal FXYD12 and NKA in the two medakas. However, different patterns of NKA activities and different interaction levels between FXYD12 and NKA were found in the kidneys of these two medakas. The results revealed that different strategies were used in the kidneys of the two medaka species upon salinity challenge. On the other hand, gene knockdown experiments demonstrated that the function of O. dancena FXYD12 allowed maintenance of a high level of NKA activity. The results of the present study indicated that the kidneys of the examined euryhaline medakas originating from brackish water and fresh water exhibited different modulatory mechanisms through which renal FXYD12 enhanced NKA activity to maintain internal homeostasis. Our findings broadened the knowledge of expression and functions of FXYD proteins, the modulators of NKA, in vertebrates.
Keywords: FXYD12, Kidney, Na+-K+-ATPase, Oryzias, Salinity.
Introduction
Euryhaline teleosts that survive in a variety of habitats must regulate their internal water and ionic concentrations to maintain their blood and tissue fluid osmolality within the range of physiological homeostasis [1-3]. Osmoregulation and ionoregulation in fish are mainly accomplished by the osmoregulatory organs, i.e., the gill, kidney, and gut [2, 3]. Among these organs, the kidneys play a crucial role in osmoregulation in euryhaline teleosts by modifying the amount of urine and maintaining ion and water balance according to environmental salinity/osmolality [1, 2]. The functions of fish kidneys are different in hyperosmotic or hypoosmotic environments, e.g., seawater (SW) or fresh water (FW), respectively. In SW, teleosts passively lose water and gain excess salt through their permeable body surface. To prevent dehydration, they ingest seawater and actively excrete small amounts of divalent ions via the kidneys, while they excrete most monovalent ions through the gills. In contrast, FW teleosts must deal with the water load and salt losses. To compensate, they absorb ions via the gill epithelia, excrete excess water and reabsorb filtered solutes in the kidneys [1-3].
In osmoregulatory organs, including the kidneys of vertebrates, such as fishes and mammals, Na+-K+-ATPase (NKA) plays important roles in osmoregulation and ion exchange by providing a driving force for many ion-transporting systems [2, 3]. NKA, a member of the P-type ATPases, is a heterodimer composed of two catalytic α-subunits, which couple ATP hydrolysis to ion transport, and two glycosylated β-subunits, responsible for the maturation and assembly of NKA [4, 5]. In euryhaline teleosts, modulation of the activity or kinetics of NKA is essential for performing its physiological functions [2, 3, 6].
Several mechanisms have been reported to participate in the regulation of NKA expression and functions [4-6]. Among the regulators of NKA, the FXYD protein (also called FXYD domain-containing ion transport regulator) family has been widely studied in mammals and elasmobranchs. The FXYD proteins are expressed in a tissue-specific manner and are characterized by the presence of a transmembrane domain and an extra-membrane FXYD motif [5, 7, 8]. The mammalian FXYD protein family consists of at least seven members (i.e., FXYD1-7), and all mammalian FXYD proteins are known to interact with NKA and affect its kinetic properties in specific ways [7-9]. In fish, shark FXYD10 was the first reported FXYD member [10]. Subsequent studies revealed the functions of FXYD10 and its interaction with NKA [11, 12]. Moreover, several FXYD proteins (i.e., FXYD2, 5-9, 11, or 12) have been identified in certain teleosts [13-18]. Relative to the mammalian FXYD proteins, FXYD9-12 are specifically found in fish [17]. Among teleostean FXYD proteins, FXYD11 and FXYD12 are distributed in a tissue-specific manner, similar to the mammalian FXYD proteins. Notably, FXYD11 is expressed prominently in the gills [14-18]. The observation of salinity-dependent mRNA and/or protein expression of branchial FXYD members suggested their physiological significance in osmoregulatory acclimation and roles in modulating the kinetic properties of NKA [13, 14, 16-21]. Although teleostean FXYD proteins have recently been identified and investigated, these studies have mainly focused on the gills of limited species.
The Indian medaka (Oryzias dancena) and the Japanese medaka (O. latipes) are euryhaline teleosts that are used as model fish, suitable for studies on salinity stress [17, 22-24]. The Indian medaka is native to brackish water (BW) environments such as river mouths and estuaries [23, 25]. Meanwhile, the Japanese medaka resides in FW environments such as ponds and paddy fields [22, 23] and is closely related to the Indian medaka [23, 26]. As the entire genome of the Japanese medaka has been sequenced and assembled, sufficient biological information can be extrapolated to the Indian medaka [26, 27], supporting the use of these species as models for studying the effects of salinity on the expression and functions of various molecules [17, 21, 24, 28-31].
Furthermore, these two species of medaka that come from different natural habitats show diverse osmoregulatory capabilities and responses (e.g., salinity tolerance, the expression patterns of branchial NKA and FXYD11) upon salinity challenge [17, 23, 24]. The differential responses between these two medaka species might result from acclimation to their natural habitats. Although the osmoregulatory capabilities of teleosts have been widely investigated, few studies have simultaneously compared the differential capabilities between two or more closely related species from diverse natural habitats. Recently, seven novel FXYD proteins were identified in different tissues/organs of these two closely related medaka species [17]. Moreover, our preliminary results revealed that fxyd12 (JX569230 and JX643983) was expressed prominently in the kidneys of both medaka species. The results suggested that FXYD12 may play crucial roles in osmoregulation via modulating NKA activity in the kidneys of the two medaka species.
It is therefore intriguing to elucidate the differences in osmoregulatory mechanisms and the relationship between these mechanisms and environmental salinities by comparing these two closely related medaka species from different natural habitats (i.e., FW vs. BW). This study aimed to compare the modulatory mechanisms of renal FXYD12 with respect to NKA expression between these two closely related medaka species from different natural habitats and to illustrate the potential role of teleostean FXYD12 upon salinity challenge. The findings of this study extended our understanding of the tissue-specific expression and functions of FXYD proteins, the regulators of NKA, in vertebrates.
Materials and Methods
Experimental animals and environments
Adult Indian medaka, identified by sequencing the 12S and 16S mitochondrial rRNA genes [32], were obtained from a local aquarium, showing a standard length of 2.5 ± 0.3 cm. Adult reddish orange Japanese medaka (HI strain), approximately 2.6 ± 0.3 cm in standard length, were inbred in the laboratory. For the experiments, the two medaka species were acclimated to FW, BW (15‰), or SW (35‰) for at least four weeks at 28 ± 1°C under a 14-h light: 10-h dark cycle [17, 24]. BW and SW were prepared from aerated dechlorinated FW (local fresh tap water) by adding standardized amounts of Instant Ocean Synthetic Sea Salt (Aquarium Systems, Mentor, OH, USA). The fish were fed a daily diet of commercial pellets provided ad libitum. In the following experiments, the fish were not fed and were anesthetized with MS-222 (100-200 mg L-1) before sampling. The protocol employed for the experimental fish was reviewed and approved by the Institutional Animal Care and Use Committee of the National Chung Hsing University (IACUC approval no. 96-48).
Probe construct used for quantitative real-time PCR
| Genes | Medaka | Primer sequence (5' to 3') | |
|---|---|---|---|
| Forward | Reverse | ||
| fxyd5 | Od | ACAGCCTGCGTGGATGAG | TAAGCGTGGTGAGCAGGAAC |
| Ol | GTTGGATGAGGAGGAAGTGG | TTGTGTTCCAGGTCGCATC | |
| fxyd6 | Od | TCACTCCTGGTATGCGTGTC | AGGCCAGTCCTCCAATTCTC |
| Ol | GTGTGCGTGTCAGCTGTTG | AGAATTGGAGGACTGGCCTT | |
| fxyd7 | Od | CTCCAAATCCAATTCCAACG | CTGGTGCATAGTTGGTGGTG |
| Ol | AACCTTGCGGACAACAGG | CGCGACAAACAAAACAACTG | |
| fxyd8 | Od | CTCCTGCACTCGCTTCAG | AGGAGGCAGAGTAGGACTGC |
| Ol | TCATTGTGTTGGTGGCATTC | ACCGATTCGCAGAGATTCA | |
| fxyd9 | Od | TGCAAGTTCAACCAGGACAA | TTGGAGCTTAGCAGTTGCAG |
| Ol | GAAGATCTGCGCTTTGGTG | ACCGCAGCAAGAATGAGG | |
| fxyd11 | Od | GGCTCGTCATTGTCTGCTTG | GGTCAGATCGCACTGCTAGA |
| Ol | CGGACTCTGTGTTGGTGAAG | ATGACGAGGCCTCCAATTC | |
| fxyd12 | Od | GGCGTTGTTGTGTTCTTGTC | CTCAGTGCAGCTCAGTCATC |
| Ol | GACAGACGAGGCAGCATGT | TCTGATCTTTGCTGGCATTG | |
| nka α | Od, Ol | GAACCGTCACCATCCTCTG | GGCTGCCTCTTCATGATGTC |
| rpl7 | Od, Ol | GTTCTGCAGCTTCTCCGTCT | GAGCTCTCGCACAGACTTCA |
| β-actin | Od, Ol | CTGGACTTCGAGCAGGAGAT | AGGAAGGAAGGCTGGAAGAG |
Od, Indian medaka; Ol, Japanese medaka; nka α, NKA α-subunit; rpl7, ribosomal protein L7.
Total RNA extraction and reverse transcription
The methods used in this study were modified from our previous studies [17, 30]. Briefly, total renal RNA samples from the two medaka species were extracted using RNA-Bee™ (Tel-Test, Friendwood, TX, USA) following the manufacturer's instructions, and genomic DNA was then eliminated using the RNA clean-up protocol of the RNAspin Mini RNA isolation kit (GE Health Care, Piscataway, NJ, USA). The concentration of the extracted RNA was measured with a NanoDrop 2000 (Thermo, Wilmington, DE, USA). For reverse transcription, cDNAs were synthesized with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA) using extracted RNA samples stored at -80°C, according to the manufacturer's instructions. The cDNA products were stored at -20°C until analysis via quantitative real-time polymerase chain reaction (PCR).
Quantitative real-time PCR
The methods used for these experiments were modified from a previous study by our group [17]. Quantitative real-time PCR was performed using the MiniOpticon real-time PCR system (Bio-Rad, Hercules, CA, USA). The PCR assays contained 8 μL of cDNA (1000× dilution), 2 μL of either 1 μM target gene primers (fxyd or nka α-subunit) or 1 μM internal control primers (β-actin for salinity-effect analyses, ribosomal protein L7 (rpl7) for other experiments), and 10 μL of 2× SYBR Green Supermix (Bio-Rad). The PCR analysis and the formula used to calculate target gene expression were described by Kang et al. [30, 31]. Probe constructs for quantitative real-time PCR were shown in Table 1. The detail information of these primers have been reported in our previous studies [17, 24, 30]. PCR products were subcloned into the pOSI-T vector (Genemark, Taipei, Taiwan) and sequenced to confirm PCR products.
Antiserum/antibody
The following primary antisera/antibodies were used in the present study: (1) FXYD12: an affinity-purified rabbit polyclonal antibody (LTK BioLaboratories, Taoyuan, Taiwan) raised against a specific peptide (NKIRRCGKPKPKPILEDD) corresponding to the C-terminal region of the cloned FXYD12 protein from the Indian medaka. The protein sequence identities between FXYD12 antibody and all FXYD proteins of the two medakas were estimated using the Bio-Edit software (data not shown). The results showed that most identities were less than 30%, except OdFXYD12 (O. dancena FXYD12; 100%), OlFXYD12 (O. latipes FXYD12; 83%), and OlFXYD7 (39%). This antibody was used for immunoblot, immunoprecipitation, and immunostaining. (2) NKA: a mouse monoclonal antibody (α5; Developmental Studies Hybridoma Bank, Iowa City, IA, USA) raised against the α-subunit of the avian NKA was employed for immunoblot, immunoprecipitation, and immunostaining. (3) Actin: a rabbit polyclonal antibody (sc-1616-R; Santa Cruz, Santa Cruz, CA, USA) against the C-terminus of human actin was applied in immunoblot assays as the loading control for adult fish. (4) RPL7: a rabbit polyclonal antibody (ab72550; Abcam, Cambridge, UK) raised against human RPL was used as a loading control for embryonic immunoblot assays.
The secondary antibodies employed for immunoblotting were horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (#0031430 or #0031460, respectively; Pierce, Rockford, IL, USA). For immunofluorescent staining, the secondary antibodies were Alexa-Fluor-488 conjugated goat anti-mouse IgG or Alexa-Fluor-546-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR, USA). Preliminary experiments of negative controls (sections stained with only primary or secondary antibodies) for all antibodies showed nonspecific staining or overstaining (data not shown).
Preparation of tissue homogenates
The methods used in this study to prepare tissue homogenates were modified from our previous studies [17, 24]. Kidneys from the two medaka species were dissected and immediately stored in a microcentrifuge tube at -80°C. Sample scrapings were suspended in a mixture of homogenization medium (SEID buffer; 300 mM sucrose, 20 mM EDTA, 100 mM imidazole and 0.1% sodium deoxycholate, pH 7.5) containing a proteinase inhibitor (#11836145001; Roche). Homogenization was performed in 2 mL microtubes with a Polytron PT1200E (Kinematica, Lucerne, Switzerland) at maximal speed for 30 sec on ice. The homogenate was then centrifuged at 5000 ×g at 4°C for 5 min. The sample supernatants were collected and processed immediately for determination of NKA activity or protein concentration measurements or stored at -80°C for immunoblotting or immunoprecipitation analysis. Protein concentrations were determined with the BCA Protein Assay (Pierce) using bovine serum albumin (Pierce) as a standard.
Immunoblotting
The immunoblotting procedures were modified from Yang et al. [17, 33]. For immunoblots of FXYD12 and the NKA α-subunit, aliquots containing 10 µg of the tissue supernatants were heated with denaturing buffer at 60 °C for 15 min, followed by electrophoresis on a 7.5% (for NKA) or 14% (for FXYD12) sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis and transformation, the blots (PVDF membranes; Millipore, Bedford, MA, USA) were pre-incubated in PBST (phosphate-buffered saline with 0.05% Tween 20) containing 5% (wt/vol) nonfat dried milk to minimize non-specific binding. Then, the blots were incubated at 4 °C overnight with NKA (α5; 2500× dilution), FXYD12 (60000× dilution), or actin (6000× dilution) in 1% bovine serum albumin (Bio Basic, Markham, Canada) and 0.05% sodium azide (Sigma, St. Louis, MO, USA) in PBST, followed by incubation with an horseradish peroxidase-conjugated secondary antibody diluted in PBST. The blots were finally developed using the ImmobilonTM Western (Millipore) and observed in a Universal hood with a cooling-CCD (charge-coupled device) camera (ChemiDoc XRS+, Bio-Rad) and the associated software (Quantity One v4.6.8, Bio-Rad). Immunoreactive bands were analyzed using Image Lab v3.0 (Bio-Rad). The results were converted to numerical values to compare the relative protein abundances of the immunoreactive bands.
The immunoblotting protocol for the negative control of FXYD12, which was used to confirm that the immunoreactivity was due to the presence of FXYD12 rather than non-specific binding, was identical to that described above except that rabbit pre-immune serum was substituted for the primary antibody.
Paraffin section preparation and immunohistochemical staining
The protocols used herein were modified from our previous studies [24, 34]. For paraffin sections, in brief, after fixation and dehydration, the tissues were embedded into paraffin blocks and then the samples were cut at 5 μm thickness. The paraffin sections were deparaffinized with xylene, rehydrated in a graded series of alcohol, and then incubated with 3% H2O2 for 10 min to inactivate endogenous peroxidase. Afterward, the slides were incubated with primary antibody (α5 or FXYD12) or PBS (negative control) followed by a commercial kit (PicTure™; Zymed, South San Francisco, CA, USA). After staining, the sections were stained with hematoxylin for histological evaluation. Finally, the sections were observed under an optical microscope (BX50, Olympus, Tokyo, Japan) equipped with the cooling-CCD camera (DP72, Olympus) using the associated software (CellSens standard v1.4, Olympus).
To identify different segments of nephron, another sets of sections were subjected to periodic acid-Schiff (PAS) staining counterstained with hematoxylin. The methods for PAS staining were described in our previous study [35]. Moreover, the characteristics for identifying the segments were based on description of previous studies [35, 36]. Briefly, the glomerulus was surrounded by the Bowman's capsule composed of capillaries, and the apical membranes of proximal tubules could be identified by the well-developed PAS-positive brush border. The distal tubule segment was composed of a single layer of epithelial cells with a centrally located nucleus and was PAS-negative at the apical side, and the collecting tubule segment was consisted of a single layer of columnar epithelial cells with basolaterally located nuclei and their lumen diameters were greater than those of proximal or distal tubules.
Cryosectioning, double immunofluorescence staining, and confocal microscopy
The methods used in this study were modified from previous studies [31, 33]. Tissues were excised and fixed with 10% neutral buffered formalin, infiltrated with OCT (optimal cutting temperature) compound (Sakura, Tissue-Tek, Torrance, CA, USA) overnight at 4°C and then mounted for cryosectioning. The tissue was cryosectioned (5 μm) using a Cryostat Microtome (CM3050S, Leica, Wetzlar, Germany) at -25°C. Sections were placed on 0.01% poly-L-lysine-coated slides (Sigma) and kept in slide boxes at -20°C.
For staining, cryosections were rinsed by PBS and then incubated with blocking buffer (5% BSA in PBS) for 30 min. After wash with PBS, cryosections were incubated with primary polyclonal antibody (FXYD12; 400× dilution in PBS) overnight at 4 °C. Subsequently, cryosections were incubated with secondary antibody (Alexa-Fluor-546 goat anti-rabbit IgG) at room temperature for 2 hrs. After the first staining, cryosections were incubated with primary monoclonal antibody (α5; 200× dilution in PBS) for 2 hrs at room temperature and then with secondary antibody (Alexa-Fluor-488 goat anti-mouse IgG) at room temperature for 2 hrs. Finally, cryosections were observed with a fluorescent microscope (BX50, Olympus) with Olympus DP72 CCD camera as described in previous paragraph or a laser scanning confocal microscope (FV1000, Olympus).
NKA activity assay
Enzyme activity was measured using the NADH-linked method [24, 33]. ADP derived from the hydrolysis of ATP by ATPase was enzymatically coupled to the oxidation of reduced NADH using lactate dehydrogenase and pyruvate kinase. The reaction was initiated by mixing 10 μL of the sample supernatant and 200 μL the reaction mixture (0.38 mM ATP, 1.50 mM phosphoenolpyruvate, 0.24 mM NADH, 2.48 U mL-1 lactate dehydrogenase, 2.70 U mL-1 pyruvate kinase, 47.3 mM NaCl, 2.63 mM MgCl2. 6H2O, 10.5 mM KCl, and 50 mM imidazole, pH 7.5) with or without 0.75 mM ouabain per well; each sample was assayed in triplicate. The 96-well plate was subsequently read every 15 sec for up to 10 min in a VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 340 nm under a temperature of 28°C. The linear rate from 2 to 8 min was determined for each pair of triplicate wells. The standard curve was determined using 10 μL of ADP per well at concentrations of 0 to 30 nmol at 340 nm at a temperature of 28°C, and the plate was read for at least 5 min after adding 200 μL of the reaction mixture (without 0.75 mM ouabain). The slope of the standard curve should be from -0.012 to -0.015. The NKA activity was calculated as the difference in the slope of ATP hydrolysis (NADH reduction) in the presence and absence of ouabain, and the activity was expressed as μmol ADP per mg protein per hour.
Co-immunoprecipitation
The methods used for co-immunoprecipitation were modified from previous studies by our group [17, 21]. Total lysates of kidneys were used in this experiment. Immunoprecipitation with the FXYD12 antibody, pre-serum (negative control), or the NKA antibody was carried out using the ImmunoCruzTM IP/WB Optima System (sc-45042 or sc-45043; Santa Cruz) according to the manufacturer's manual. After elution, the samples were stored at 4°C until use. To confirm the interaction between FXYD12 and the NKA α-subunit, the obtained immunoprecipitation solutions were analyzed by immunoblotting as described above. To investigate the effects of salinity on the interaction between FXYD12 and NKA α-subunit in the two medaka species, immunoprecipitation solutions containing the NKA α-subunit were obtained from total renal lysates of FW-, BW-, or SW-acclimated individuals and analyzed via immunoblotting to detect the NKA α-subunit or FXYD12. The following immunoblotting protocol was employed as described above, except that anti-NKA (α5) was substituted for the loading control. The results were converted to numeric values (normalized relative to NKA expression) to compare the relative protein abundance of the immunoreactive bands.
Morpholino (MO) oligonucleotide knockdown in the Indian medaka
MOs of Odfxyd12 anti-sense were designed and manufactured by Gene Tools (Philomath, OR, USA). OdFXYD12-MO (5'-AGAAAACTGAGCTGCTGAAGGCTCC-3') acted as a translational blocker. For negative control experiments, SC (standard control oligo; 5'-CCTCTTACCTCAGTTACAATTTATA-3'; Gene Tools) was used. These oligonucleotides were dissolved in RNase-free water to obtain 5 mM injection stocks. To confirm the safety and efficiency of the MOs, various injection dosages of the MOs were tested, and a dosage of 1 μM per embryo was selected for subsequent injections. Using this dosage, neither significant mortality nor abnormal embryos were found.
The methods used in this study were modified from our previous study [31]. Fertilized eggs of BW-acclimated Indian medaka at the one- or two-cell stage were collected and then immersed in balanced salt solution (111.1 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2 · 2H2O, 0.8 mM MgSO4- · 7H2O, 5% NaHCO3, 0.01% phenol red) at a low temperature (16-18°C) to extend the developmental period into the four-cell stage. The solutions containing OdFXYD12-MO or SC were injected (approximately 2-5 nL) into embryos at the one- or two-cell stage using a microinjection system consisted of an Olympus SZ60 stereomicroscope (Tokyo, Japan), an air-pressure driven microinjector (IM-300; Narishige, Tokyo, Japan), and a three-axis micromanipulator (M-152, Narishige). The injected embryos were incubated in balanced salt solution at 28 ± 1°C, and the media were replaced every day. To confirm the protein expression and localization of FXYD12 and NKA α-subunit as well as NKA activity in the developmental process of Indian medaka after fertilization, their expression profiles were investigated (data not shown). During embryonic development, expression of these proteins was significantly enhanced at two days post fertilization (2 dpf) and at all later developmental stages. Thus, these embryos at 2 dpf were sampled for subsequent analyses.
Preparation of embryonic homogenates and crude membrane fractions
The methods used in this experiment were similar to the methods employed to obtain tissue homogenates from the adults, with slight modifications. Briefly, each total protein sample was assembled from three eggs stored in a microcentrifuge tube at -80°C before use. After homogenization and centrifugation, the sample supernatants were processed immediately for determination of NKA activity and protein concentrations or stored at -80°C for embryonic FXYD12 immunoblotting.
For immunoblotting analysis of the embryonic NKA α-subunit, embryonic crude membrane fractions were also prepared. The methods used in this study were modified from previous studies [16, 37]. Briefly, each total protein sample was assembled from 10 eggs stored in a microcentrifuge tube at -80°C before use. After homogenization, centrifugation (13000 ×g, 4°C for 10 min) was first performed to remove debris, nuclei and lysosomes. The remaining supernatant was then centrifuged at 20800 ×g at 4°C for 1 hr. The resulting pellet was resuspended in SEID buffer and stored at -80°C until use. The pelleted fraction is expected to contain large fragments from the plasma membrane (both apical and basolateral), Golgi body, and endoplasmic reticulum [38]. Thus, this fraction was referred to as the crude membrane fraction. The sample fractions were collected and used to perform measurements of protein concentrations and NKA immunoblotting.
Analyses of MO effects
To investigate the effects of gene knockdown of the FXYD12 protein, the changes in the protein abundance of FXYD12 and NKA α-subunit as well as NKA activity in the 2 dpf embryos of Indian medaka were determined. The methods used to assay NKA activity in this experiment were previously described, and the immunoblot protocols were similar with slight modifications. Briefly, an aliquot containing 5 or 40 µg (for NKA or FXYD12, respectively) of the embryonic supernatant was added to sample buffer and heated at 60ºC for 15 min, followed by electrophoresis on a 9% (for NKA) or 14% (for FXYD12) sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis, transformation, and pre-incubation, the blots were incubated at 4°C overnight with NKA (α5; 5000× dilution), FXYD12 (10000× dilution), or RPL7 (10000× dilution) in 1% bovine serum albumin and 0.05% sodium azide in PBST and subsequently with the appropriate secondary antibody diluted in PBST. The blots were observed as described above. The results were converted to numerical values to compare the relative protein abundances of the immunoreactive bands.
Statistical analysis
The values are expressed as the means ± SEM (standard error of the mean). The results were compared via one-way ANOVA with Tukey's pairwise method using SPSS 12.0 software (SPSS Inc., Chicago, IL, USA), and P<0.05 was set as the significance level.
Results
Renal fxyd mRNA levels in Indian medaka and Japanese medaka
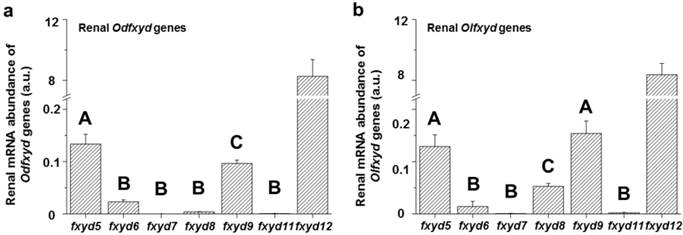
The expression patterns of the renal fxyd genes were similar between the Indian medaka (Odfxyd) and the Japanese medaka (Olfxyd) (Fig. 1). In the kidney, the level of fxyd12 mRNA was the highest among all of the detected fxyd genes. The mRNA levels of fxyd5 or/and fxyd9 were the second highest in both medaka species. Moreover, the renal mRNA levels of fxyd12 were over 50-fold higher than those of fxyd5 and fxyd9 in both Indian medaka and Japanese medaka. Conversely, the mRNA levels of fxyd7 and fxyd11 were low in the kidneys of both medaka species.
Immunodetection of FXYD12 protein
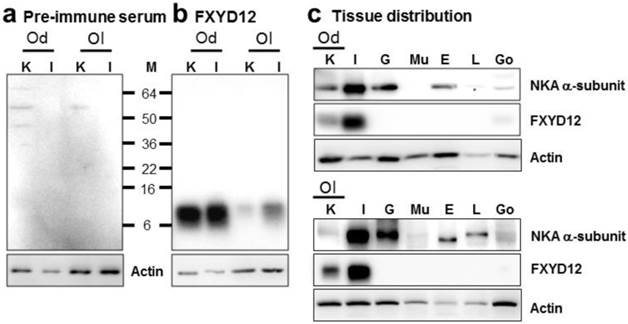
To investigate the expression pattern of FXYD12, an antiserum was made against a specific peptide corresponding to the C-terminal region of the Indian medaka FXYD12. Comparing with the results of the negative control in which the primary antibody was replaced by the pre-immune serum from the pre-immunoreactive rabbit (Fig. 2a), the results of immunoblots showed a single immunoreactive band of FXYD12 in both kidney and intestine with a molecular mass at approximately 10 kDa in these two medaka species (Fig. 2b). The results showed that the FXYD12 antibody can be successfully applied to detect the FXYD12 proteins of both medakas. On the other hand, the results of tissue distribution revealed that expressions of FXYD12 protein were found mainly in the kidney and intestine of both Indian medaka and Japanese medaka (Fig. 2c). In both medaka species, moreover, the NKA α-subunit expressions were found in various tissues including the kidney (Fig. 2c).
Renal FXYD12 and NKA distribution of the two medaka species
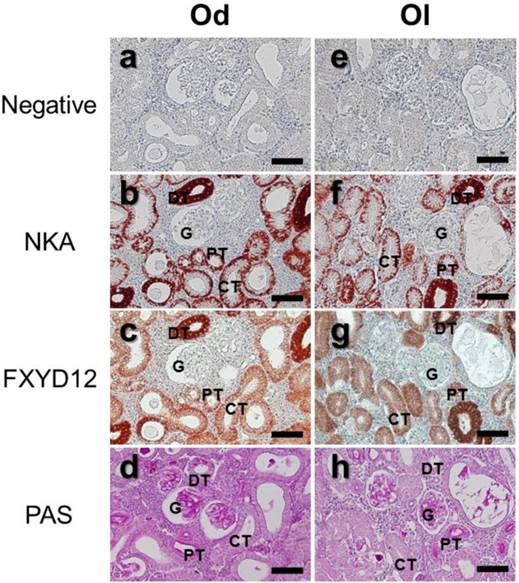
The locations of FXYD12 and NKA were identical in kidneys between two medaka species (Fig. 3). According to the results of PAS staining (Fig. 3d, h), the nephron of the two medakas both included the glomerulus, proximal tubules, distal tubules, and collecting tubules. Thus, immunohistochemical staining revealed that both NKA and FXYD12 were distributed in the proximal tubule, distal tubule, as well as collecting tubules, but not in the glomeruli of the Indian medaka (Fig. 3b, c) and the Japanese medaka (Fig. 3f, g). Negative control without application of antibodies revealed no immunoreaction in the renal paraffin sections of both species (Fig. 3a, e). Confocal 3D micrographs confirmed the colocalization of FXYD12 and NKA in the BW-acclimated Indian medaka kidneys (Fig. 4). Further double immunofluorescence staining showed that FXYD12 was localized in the NKA-immunoreactive (NKA-IR) cells in renal tubules of all salinity groups of the two medaka species (Figs. 5, 6).
Comparisons of renal fxyd mRNA abundance in the Indian medaka (Od; a) and the Japanese medaka (Ol; b). The results combined the data of all salinity groups. The values are means ± SEM (N=12). Different letters indicate significant differences among fxyd genes, excluding fxyd12 (P<0.05). A.u., arbitrary units.
Representative immunoblots from the kidney and intestine of the brackish water-acclimated Indian medaka (Od) and fresh water-acclimated Japanese medaka (Ol) for FXYD12 and the NKA α-subunit (NKA). (a) Pre-immune serum was used as the negative control for FXYD12, which revealed no immunoreactive band in the kidney (K) or intestine (I). (b) The immunoblots showed a single immunoreactive band for the FXYD12 antibody, with a molecular mass of approximately 10 kDa. (c) The immunoblots for different tissues showed that NKA occurred in the kidneys (K) and intestines (I) as well as the gills (G), eyes (E), livers (L), and gonads (Go) while FXYD12 was detected mainly in the kidneys and intestines. Actin was used as an internal control for the immunoblots. M, marker (kDa); Mu, muscle.
Distribution of NKA α-subunit (NKA) and FXYD12 in paraffin sections of kidneys of the brackish water-acclimated Indian medaka (Od; a-d) and fresh water-acclimated Japanese medaka (Ol; e-h). Immunosignals of NKA (b, f) and FXYD12 (c, g) were both detected in the nephron epithelial cells, comparing with the negative control (a, e). Different segments of the nephron were identified by periodic acid-Schiff (PAS) staining counterstained with hematoxylin (d, h). G, glomeruli; PT, proximal tubule; DT, distal tubule; CT, collecting tubule. Scale bar: 50 μm.
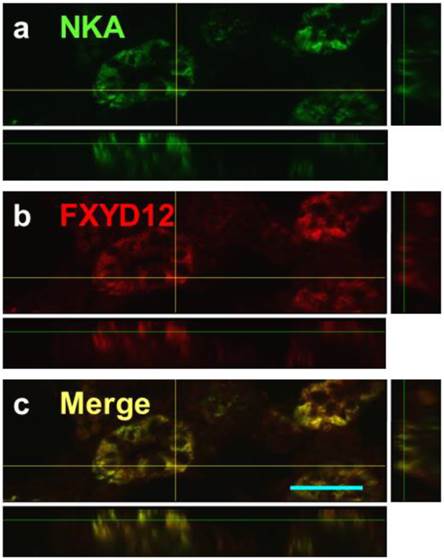
Confocal 3D micrographs of double immunofluorescence staining of NKA α-subunit (NKA; green; a) and FXYD12 (red; b) in the cryosection of the brackish water-acclimated Indian medaka kidneys. The merged image (yellow; c) revealed that FXYD12 colocalized to NKA-immunoreactive cells of the proximal tubule. Scale bar: 20 μm.
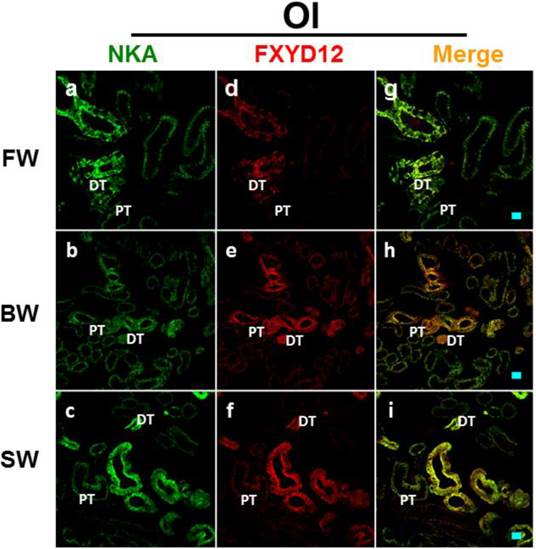
Double immunofluorescence staining of NKA α-subunit (NKA; green; a-c) and FXYD12 (red; d-f) in renal cryosections of the Indian medaka (Od). The merged images (yellow; g-i) revealed that FXYD12 colocalized to the NKA-immunoreactive cells in the fresh water- (FW-; a, d, g), brackish water- (BW-; b, e, h), and seawater- (SW-; c, f, i) acclimated fish. PT, proximal tubule; DT, distal tubule. Scale bar: 20 μm.

Double immunofluorescence staining of NKA α-subunit (NKA; green; a-c) and FXYD12 (red; d-f) in renal cryosections of the Japanese medaka (Ol). The merged images (yellow; g-i) revealed that FXYD12 colocalized to the NKA-immunoreactive cells in the fresh water- (FW-; a, d, g), brackish water- (BW-; b, e, h), and seawater- (SW-; c, f, i) acclimated fish. PT, proximal tubule; DT, distal tubule. Scale bar: 20 μm.
Effects of salinity on the expression of renal FXYD12 and NKA in the medakas
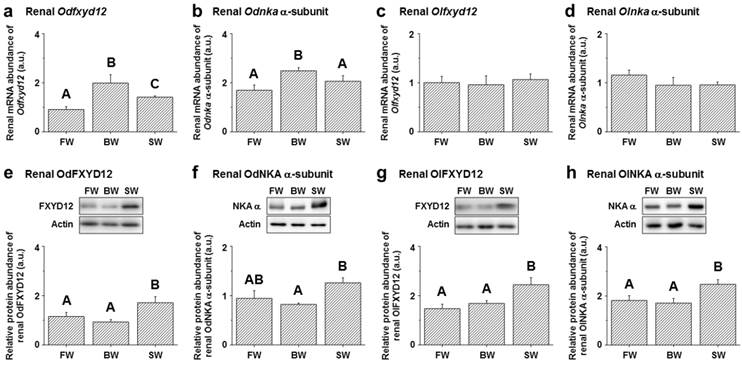
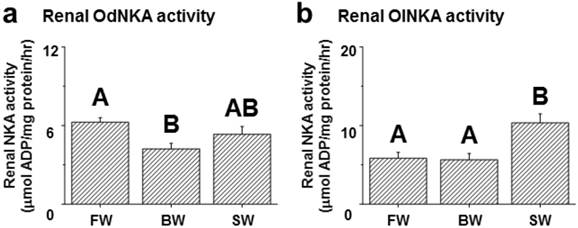
Among the assessed mRNA levels, similar expression patterns were found between fxyd12 and the nka α-subunit in the kidneys of the Indian medaka and Japanese medaka (Fig. 7a-d). The highest expression of Odfxyd12 and Odnka was found in the kidneys of the BW-acclimated Indian medaka (Fig. 7a, b). Moreover, the results showed that the expression patterns of Olfxyd12 and Olnka in the Japanese medaka were not significantly different among various salinity groups (Fig. 7c, d). On the other hand, the expression patterns concerning renal FXYD12 and NKA α-subunit protein abundance were similar in the two medaka species, with higher expression being found in the SW-acclimated individuals than in the FW- and BW-acclimated groups (Fig. 7e-h). The specific activity of NKA in the kidneys of the FW-acclimated Indian medaka was significantly higher than in the BW-acclimated fish (Fig. 8a). A slightly higher NKA activity was found in the SW-acclimated Indian medaka compared with the BW group, although there was no significant different (Fig. 8a). In addition, the highest levels of renal NKA activity were found in the SW-acclimated Japanese medaka compared with other groups (Fig. 8b).
Effects of salinity on mRNA levels (a-d) and protein abundance (e-h) of renal FXYD12 and NKA α-subunit (NKA α) in the Indian medaka (Od) and the Japanese medaka (Ol). The values are means ± SEM (N=5-10). Dissimilar letters indicate significant differences among the various salinity groups (P<0.05). Actin was used as an internal control for the immunoblots. FW, fresh water; BW, brackish water; SW, seawater; a.u., arbitrary units.
Effects of salinity on renal NKA activity in the Indian medaka (Od; a) and the Japanese medaka (Ol; b). The values are means ± SEM (N=7-12). Dissimilar letters indicate significant differences among the various salinity groups (P<0.05). FW, fresh water; BW, brackish water; SW, seawater.
Association of FXYD12 with NKA in the kidneys of the two medaka species
The interaction between renal FXYD12 and NKA in the two medaka species was investigated via immunoprecipitation and immunoblotting (Fig. 9). The results showed that immunobands appeared at 10 and 100 kDa for FXYD12 and NKA, respectively, after immunoprecipitation (Fig. 9). Meanwhile, the positive and negative controls (lane 1 and lane 2 of Fig. 9) demonstrated the immunoprecipitation efficiency.
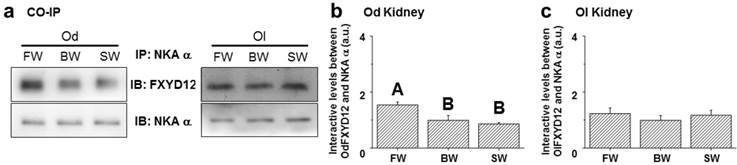
To examine the effects of salinity on the interaction between renal FXYD12 and NKA, immunoprecipitated NKA α-subunit (α5) was prepared in the complexes of immunoprecipitation for NKA and FXYD12 using immunoblot analysis (Fig. 10a). In the Indian medaka, the NKA protein abundance was found to be constant among different salinity groups, while the highest level of FXYD12 protein was found in the FW-acclimated individuals (Fig. 10b). The results illustrated that the interaction level between FXYD12 and NKA was increased in the kidneys of the FW-acclimated Indian medaka. In the kidneys of the Japanese medaka, however, there were no significant differences in the associations among all groups (Fig. 10c).
Effects of gene knockdown of the OdFXYD12 protein in Indian medaka embryos
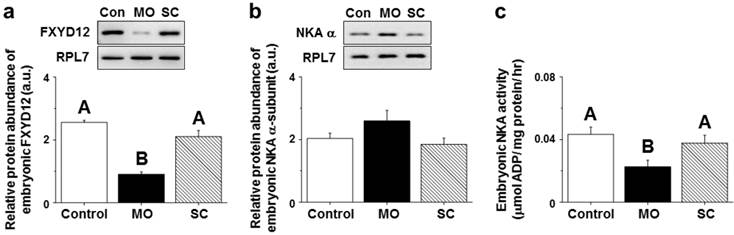
After injecting MOs (morpholino oligonucleotides) targeting the OdFXYD12 protein, the protein abundance of embryonic OdFXYD12 was significantly decreased compared with the untreated (control, Con) and SC (standard control oligo) groups (Fig. 11a). For the protein abundance of the embryonic NKA α-subunit, although there was no statistically significant difference among the three groups (P=0.169), a slightly higher level was found in the MO group compared with the untreated and SC groups (Fig. 11b). On the other hand, decreased embryonic NKA activity was found in the OdFXYD12-MO embryos compared with either the untreated or SC group (Fig. 11c). In addition, there was no significant difference between the untreated and SC groups in terms of NKA activity or the protein abundance of OdFXYD12 and NKA α-subunit (Fig. 11).
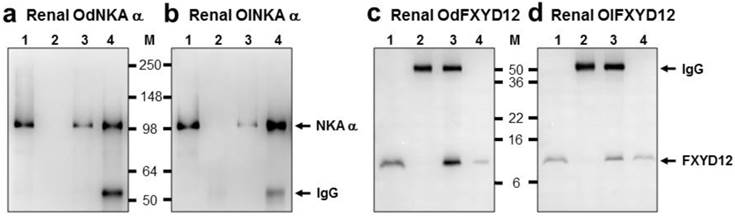
Co-immunoprecipitation of FXYD12 and NKA α-subunit (NKA α) in the kidneys of the Indian medaka (Od; a, c) and the Japanese medaka (Ol; b, d). Immunoreactive bands of NKA α (a, b) or FXYD12 (c, d) were detected at 100 or 10 kDa, respectively. Lane 1, positive control (total renal lysates); lane 2, negative immunoblot control using pre-immune serum for immunoprecipitation; lanes 3 and 4, experimental group using antibodies (FXYD12 and NKA α, respectively) for immunoprecipitation. The 55 kDa bands in lanes 2-4 are the IgG heavy chain of the FXYD12 or NKA α antibody. M, marker (kDa).
Effects of salinity on the interaction between FXYD12 and NKA α-subunit (NKA α) in the Indian medaka (Od) and the Japanese medaka (Ol). Immunoprecipitation (IP) of NKA α was performed using renal total lysates from fresh water- (FW), brackish water- (BW), or seawater- (SW) acclimated medaka, and the complexes were then analyzed in immunoblots (IB) for NKA α or FXYD12. The values are means ± SEM (N=5). Dissimilar letters indicate significant differences among various salinity groups (P<0.05). A.u., arbitrary units.
Gene knockdown effects of the FXYD12 protein on the protein abundance of FXYD12 (a) and NKA α-subunit (NKA α; b) as well as NKA activity (c) in the Indian medaka embryos at two days post-fertilization. PRL7 was used as an internal control for immunoblotting. The values are means ± SEM (N=3 or 6 for NKA immunoblots or others, respectively). Dissimilar letters indicate significant differences among the various salinity groups (P<0.05). MO, embryos microinjected with OdFXYD12-MO; SC, embryos microinjected with the standard control oligo; Con, control (untreated embryos); a.u., arbitrary units.
Discussion
FXYD proteins belong to a family of small-membrane proteins that associate with and play a role as modulators of NKA [5, 7, 8]. In mammals and elasmobranchs, significant effects of FXYD proteins (FXYD1-7 and FXYD10) in altering NKA kinetic properties have been demonstrated using various expression systems [5, 7-9]. Moreover, in teleosts, renal NKA plays a crucial osmoregulatory role in maintaining intracellular homeostasis [1, 2]. Therefore, the present study investigated the salinity-dependent expression and potential function of FXYD12 in the kidneys of two closely related congeneric fish species (the Indian medaka and Japanese medaka) with different natural habitats to illustrate the modulatory mechanisms of the teleostean FXYD12 protein to shed more light on the molecular mechanisms underlying the regulation of NKA activity in euryhaline teleosts.
FXYD12, the major member of FXYD protein family in medaka kidneys
The results in this study revealed that in both medaka species, the mRNA levels of fxyd12 were the most abundant among the examined fxyd genes, indicating that FXYD12 is the major member of the FXYD family in kidneys of these two medaka species. In protein tissue distribution, moreover, the FXYD12 was found mainly in the kidney and intestine of both medaka species. The results were not only consistent with those of fxyd12 gene in these two medaka species [17] but also similar to the distribution profiles of fxyd12 mRNA in the Atlantic salmon (Salmo salar)[14], zebrafish (Danio rerio)[15], and spotted scat (Scatophagus argus)[18]. On the other hand, the results of immunostainings showed that FXYD12 was colocalized to the basolateral membrane of NKA-IR cells in the proximal, distal, and collecting tubules of both medaka species. Although no study reported the localization of FXYD12 to date, NKA distribution has been described in previous papers in the other euryhaline teleosts [35, 36, 39, 40]. These results of NKA localization were similar among these two medakas and other teleosts. Marshall and Grosell [2] indicated that the highest expression of NKA was found in the distal tubule responsible for reabsorption of monovalent ions in fish adapted to various environments. In mammals, furthermore, several FXYD proteins (e.g., FXYD2, FXYD4, and FXYD5) have mainly been detected in the kidneys and localized to the basolateral membrane of the nephron tubules [7, 8, 41]. Therefore, these results revealed that the FXYD proteins mainly expressed in the kidneys were different between teleosts and mammals. These results also implied that FXYD12 played a crucial role in NKA-IR cells of teleostean kidneys, similar to the mammalian renal FXYD proteins.
Similar salinity-dependent protein expression of FXYD12 and NKA in medaka kidneys
Upon salinity challenge, although the mRNA expression profiles of fxyd12 and nka were different between the Indian medaka and Japanese medaka, the patterns of abundance were similar among all of the detected proteins (OdFXYD12, OdNKA, OlFXYD12, and OlNKA) in this study. In addition, the protein levels of FXYD12 and NKA showed different patterns compared with their mRNA expression. These results of diverse expression were also found in branchial NKA expression of these two medaka species [24] as well as in branchial FXYD11 expression of the Atlantic salmon [19]. The inconsistency on patterns between mRNA and protein levels may be due to translation efficiency, protein stability, post-transcriptional and/or post-translational changes in protein expression [19]. In this study, the highest expression of the NKA and FXYD12 proteins was found in the kidneys of SW-acclimated Indian medaka and Japanese medaka. To date, renal FXYD12 expression (mRNA level) has only been reported in the Atlantic salmon [14] and spotted scat [18]. Unlike the medaka species examined in this work, in the Atlantic salmon and spotted scat, the expression of renal fxyd12 mRNA was higher in FW fish than in the SW group [14, 18]. On the other hand, in either mRNA or protein levels, expression results showed a positive correlation between FXYD12 and NKA in the kidneys of these two medaka species. A similar correlation was also found between FXYD11 and NKA in the gills of the Atlantic salmon and Indian medaka, revealing the possibility that FXYD11 may increase NKA activity [17, 19, 21].
Different renal NKA activity profiles between the two medaka species
In this study, the expression patterns of renal NKA activity were found to be different between the Indian medaka and Japanese medaka. Most studies on renal NKA in fish have focused on NKA activity, while few articles have reported the mRNA levels and/or protein abundance of NKA [35, 39, 42]. Previous studies have identified three salinity-related patterns of teleostean renal NKA activity: (1) a rise in FW-acclimated individuals [16, 35, 39, 42]; (2) an increase with environmental salinities [43, 44]; and (3) an absence of significant differences among diverse salinity groups [45, 46]. In this study, renal NKA activity in the Indian medaka increased in FW, while that in the Japanese medaka increased in SW. These results revealed that the expression profiles of NKA activity following environmental challenge varied between these teleosts. In FW (i.e., hypoosmotic environment), the major role of the teleostean kidney is to excrete excess water and reabsorb filtered solutes. In SW teleosts, however, the primary function of the kidney is to reduce the reabsorption of solutes and produce small volumes of blood-isotonic urine containing divalent ions, particularly Mg2+ and SO42- [1-3]. In both cases, renal NKA activity plays a crucial role in providing the driving force for secondary ion transporters to maintain homeostasis [1, 2]. Hence, in the Indian medaka, renal NKA might provide a greater driving force for increasing ion reabsorption when the fish survived in hypoosmotic FW. On the contrary, when the fish are confronted with dehydration and salt inflow, renal NKA might provide a greater driving force to increase water reabsorption and/or ion secretion in the Japanese medaka.
Diverse modulatory mechanisms of renal FXYD12 with NKA
The osmoregulatory capabilities of teleosts may be correlated with their evolutionary histories or natural habitats [6, 23, 47]. By comparing these two closely related species of medaka from different natural habitats, the results revealed that BW-residing Indian medaka and FW-residing Japanese medaka exhibited lower requirements for renal NKA activity (i.e., osmoregulatory force) in their natural habitats. Upon salinity challenge, lower levels (mRNA, protein, and/or activity) of branchial NKA and FXYD11 were also found in the gills of medakas when they survived in their natural habitats (i.e., in the BW-acclimated Indian medaka and FW-acclimated Japanese medaka)[17, 24]. Thus, different expression patterns of renal FXYD12 and NKA observed between these two medaka species may be related to their natural habitats. Interestingly, the expression patterns of renal proteins (both NKA and FXYD12) and NKA activity were found to be different between these two medaka species in this study. In the kidneys of the Japanese medaka, the patterns of NKA activity were consistent with the protein expression of both NKA and FXYD12. In contrast, in the Indian medaka kidneys, the highest NKA activity was found in FW-acclimated fish, differing from the pattern found for the protein abundance of NKA and FXYD12. These results revealed that the modulatory mechanisms may be different between the kidneys of these two medaka species.
Various factors such as interaction levels, differential phosphorylation, and hormones that play roles in the modulatory mechanisms of FXYD proteins and NKA have been illustrated in mammalian studies [5, 7, 48]. Among these factors, FXYD proteins have been reported to modulate NKA expression via their interaction [8, 12, 49], and the interaction level is a key factor in the modulatory mechanisms of FXYD proteins and NKA [48, 50-52]. In this study, the interactions between FXYD12 and NKA in the kidneys of the Indian medaka and Japanese medaka were demonstrated, and the effects of salinity on the interactions were different between these two medaka species. There was no significant difference among all of the investigated groups of the Japanese medaka, whereas an enhanced interaction level was found in the FW-acclimated Indian medaka. The increased interaction revealed that the association of FXYD and NKA was more efficient [48, 51, 52]. Therefore, renal FXYD12 might modulate NKA expression in the Indian medaka via alteration of the interaction levels between these proteins when salinity changes occur in the environment. Taken together, these results suggested that the kidneys of the Indian medaka and Japanese medaka followed different strategies via modulating the expression of FXYD12 and NKA and/or their association to maintain internal homeostasis in environments with various salinities.
The potential role of renal FXYD12 in the Indian medaka
The effects of FXYD proteins on NKA are diverse. In mammals, FXYD1-3 and FXYD7 inhibits the activity of NKA via decreasing its affinity for Na+ and/or K+ [51-54], while FXYD4 and FXYD5 stimulate NKA activity by increasing Vmax and/or the affinity for Na+ [41, 55]. On the other hand, shark FXYD10 inhibits the Vmax of NKA to decrease its activity [10, 11]. In teleosts, Saito et al. [15] first reported the analysis of fxyd11-MO zebrafish to explain the function of zebrafish FXYD11 and found an increased number of NKA-IR cells in the larval yolk sac skin to compensate for the functional impairment of NKA. FXYD functions in some teleosts have also been inferred based on the results of sequence similarities among FXYD proteins or similar expression patterns (mRNA/protein) between NKA and FXYD proteins, especially FXYD11 [16, 17, 19-21]. These studies have indicated that teleostean FXYD11 might enhance NKA activity in the gills [15, 17, 19, 21].
The use of antisense MOs, an effective gene knockdown technique, is a common and reliable approach for estimating the in vivo functions of target genes using zebrafish and Japanese medaka embryos [27, 56]. In fish embryos and newly hatched larvae, the osmoregulatory organs including the kidney are not yet developed or fully functional. However, the fish embryos and newly hatched larvae are able to maintain hydromineral balance via ionocytes (i.e., NKA-IR cells) in the epithelium of their yolk-sacs and trunks [15, 31, 57]. Our preliminary results revealed that, in 2 dpf embryos of the Indian medaka, most FXYD12 signals were found in NKA-IR cells on the yolk-sac epithelium (data not shown), like the distribution of FXYD11 in the yolk-sac skin of 2 dpf zebrafish larval [15]. Thus, the results showed that the FXYD12 interacted with NKA in NKA-IR cells (i.e., ionocytes) of yolk-sac epithelium of 2 dpf embryos. In addition, in a previous study, we further developed and applied the MO technique in Indian medaka embryos to reduce the translation level of target genes for functional analysis of gene products [31]. In this study, the function of FXYD12 was investigated using OdFXYD12-MO in Indian medaka embryos. Following MO microinjection, OdFXYD12 protein abundance and NKA activity were significantly decreased. These results revealed that a lack of, or down-regulated OdFXYD12 expression caused a decrease of NKA activity, while NKA protein abundance was not significantly changed. Therefore, OdFXYD12 is able to maintain a high level of NKA activity. The present study demonstrated that FXYD12 plays a crucial role in the kidneys of the medaka via maintaining (or enhancing) a high level of NKA activity for osmoregulation/ionoregulation. Taken together, in kidneys of the FW-acclimated Indian medaka in this study, the high level of NKA activity may be due to their higher interaction between NKA and FXYD12 (the NKA activity enhancer).
Conclusions
This study revealed differences in the modulatory mechanisms of renal FXYD12 relevant to NKA between two closely related euryhaline fish (the Indian medaka and Japanese medaka) upon salinity challenges. The results of immunoprecipitation and immunostainings demonstrated that FXYD12 interacted with NKA in NKA-IR cells of renal tubules of these two medakas. By the gene knockdown experiments (morpholino), the NKA activity was reduced when the FXYD12 was disappeared in medaka embryos. The results also suggested that FXYD12 was able to maintain a high level of NKA activity. Additionally, the kidneys of the Indian medaka and Japanese medaka showed different mechanisms underlying the effects of FXYD12 on NKA expression (e.g., interaction levels and others) to maintain the water and ion balance. Through localization, immunoprecipitation, and morpholino experiments, this study illustrated the modulatory mechanism and functions of the teleost FXYD12 protein and extended our understanding in vertebrates' FXYD proteins.
Abbreviations
BW: brackish water; FW: freshwater/fresh water; dpf: day(s) post fertilization; MO: morpholino oligonucleotide; NKA: Na+-K+-ATPase; NKA-IR cells: NKA-immunoreactive cells; NKA α: NKA α-subunit; Od: Indian medaka (O. dancena); Ol: Japanese medaka (O. latipes); PAS staining: periodic acid-Schiff staining; PBS: phosphate-buffered saline; PBST: PBS with 0.05% Tween 20; PCR: polymerase chain reaction; RPL7: ribosomal protein L7; SC: standard control oligo; SW: seawater.
Acknowledgements
The monoclonal antibody against the Na+-K+-ATPase α-subunit (α5) was purchased from the Developmental Studies Hybridoma Bank maintained by the Department of Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, MD 21205, and the Department of Biological Sciences, University of Iowa, Iowa City, IA 52242, under Contract N01-HD-6-2915, NICHD, USA. This study was supported by grants from the Academia Sinica of Taiwan (AS-98-TP-B08). The work was also supported in part by a grant from the ATU plan of the Ministry of Education, Taiwan (CC-97118).
Author contributions
WKY, CHL, and THL designed the project. WKY, ADH, and CKK performed the experiments. CKK contributed new reagents/analytic tools. WKY and ADH analyzed the data and wrote the paper. All authors have read and approved the final manuscript.
Conflict of interest
The authors have no competing interests to declare.
References
1. Perry SF, Shahsavarani A, Georgalis T, Bayaa M, Furimsky M, Thomas SLY. Channels, pumps, and exchangers in the gill and kidney of freshwater fishes: their role in ionic and acid-base regulation. J Exp Zool A. 2003;300:53-62
2. Marshall WS, Grosell M. Ion transport, osmoregulation, and acid-base balance. In: (ed.) Evans DH, Claiborne JB. The Physiology of Fishes. Boca Raton: CRC Press. 2006:177-230
3. Evans DH. Teleost fish osmoregulation: what have we learned since August Krogh, Homer Smith, and Ancel Keys. Am J Physiol Regul Integr Comp Physiol. 2008;295:R704-R13
4. Scheiner-Bobis G. The sodium pump. Eur J Biochem. 2002;269:2424-33
5. Crambert G, Geering K. FXYD Proteins: new tissue-specific regulators of the ubiquitous Na,K-ATPase. Sci STKE. 2003;2003:re1
6. Hwang PP, Lee TH. New insights into fish ion regulation and mitochondrion-rich cells. Comp Biochem Physiol A. 2007;148:479-97
7. Garty H, Karlish SJD. Role of FXYD proteins in ion transport. Annu Rev Physiol. 2006;68:431-59
8. Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290:F241-F50
9. Geering K. Function of FXYD proteins, regulators of Na, K-ATPase. J Bioenerg Biomembr. 2005;37:387-92
10. Mahmmoud YA, Vorum H, Cornelius F. Identification of a phospholemman-like protein from shark rectal glands. Evidence for indirect regulation of Na, K-ATPase by protein kinase C via a novel member of the FXYD family. J Biol Chem. 2000;275:35969-77
11. Mahmmoud YA, Cramb G, Maunsbach AB, Cutler CP, Meischke L, Cornelius F. Regulation of Na,K-ATPase by PLMS, the phospholemman-like protein from shark: molecular cloning, sequence, expression, cellular distribution, and functional effects of PLMS. J Biol Chem. 2003;278:37427-38
12. Mahmmoud YA, Vorum H, Cornelius F. Interaction of FXYD10 (PLMS) with Na, K-ATPase from shark rectal glands: close proximity of Cys74 of FXYD10 to Cys254 in the a domain of the α-subunit revealed by intermolecular thiol cross-linking. J Biol Chem. 2005;280:27776-82
13. Wang PJ, Lin CH, Hwang HH, Lee TH. Branchial FXYD protein expression in response to salinity change and its interaction with Na+/K+-ATPase of the euryhaline teleost Tetraodon nigroviridis. J Exp Biol. 2008;211:3750-8
14. Tipsmark CK. Identification of FXYD protein genes in a teleost: tissue-specific expression and response to salinity change. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1367-R78
15. Saito K, Nakamura N, Ito Y, Hoshijima K, Esaki M, Zhao B. et al. Identification of zebrafish Fxyd11a protein that is highly expressed in ion-transporting epithelium of the gill and skin and its possible role in ion homeostasis. Front Physiol. 2010;1:129
16. Tang CH, Lai DY, Lee TH. Effects of salinity acclimation on Na+/K+-ATPase responses and FXYD11 expression in the gills and kidneys of the Japanese eel (Anguilla japonica). Comp Biochem Physiol A. 2012;163:302-10
17. Yang WK, Kang CK, Chang CH, Hsu AD, Lee TH, Hwang PP. Expression profiles of branchial FXYD proteins in the brackish medaka Oryzias dancena: a potential saltwater fish model for studies of osmoregulation. PLoS ONE. 2013;8:e55470
18. Hu P, Li S, Zhong Y, Mu X, Gui L, Zhang J. Identification of fxyd genes from the spotted scat (Scatophagus argus): molecular cloning, tissue-specific expression, and response to acute hyposaline stress. Comp Biochem Physiol B. 2014;174:15-22
19. Tipsmark CK, Mahmmoud YA, Borski RJ, Madsen SS. FXYD-11 associates with Na+-K+-ATPase in the gill of Atlantic salmon: regulation and localization in relation to changed ion-regulatory status. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1212-R23
20. Tipsmark CK, Breves JP, Seale AP, Lerner DT, Hirano T, Grau EG. Switching of Na+, K+-ATPase isoforms by salinity and prolactin in the gill of a cichlid fish. J Endocrinol. 2011;209:237-44
21. Chang CH, Yang WK, Lin CH, Kang CK, Tang CH, Lee TH. FXYD11 mediated modulation of Na+/K+-ATPase activity in gills of the brackish medaka (Oryzias dancena) when transferred to hypoosmotic or hyperosmotic environments. Comp Biochem Physiol A. 2016;194:19-26
22. Sakamoto T, Kozaka T, Takahashi A, Kawauchi H, Ando M. Medaka (Oryzias latipes) as a model for hypoosmoregulation of euryhaline fishes. Aquaculture. 2001;193:347-54
23. Inoue K, Takei Y. Asian medaka fishes offer new models for studying mechanisms of seawater adaptation. Comp Biochem Physiol B. 2003;136:635-45
24. Kang CK, Tsai SC, Lee TH, Hwang PP. Differential expression of branchial Na+/K+-ATPase of two medaka species, Oryzias latipes and Oryzias dancena, with different salinitytolerances acclimated to fresh water, brackish water and seawater. Comp Biochem Physiol A. 2008;151:566-75
25. Roberts TR. Systematic observations on tropical Asian medakas or ricefishes of the genus Oryzias, with descriptions of four new species. Ichthyol Res. 1998;45:213-24
26. Sasado T, Tanaka M, Kobayashi K, Sato T, Sakaizumi M, Naruse K. The National BioResource Project Medaka (NBRP Medaka): an integrated bioresource for biological and biomedical sciences. Exp Anim. 2010;59:13-23
27. Wittbrodt J, Shima A, Schartl M. Medaka - a model organism from the far east. Nat Rev Genet. 2002;3:53-64
28. Kang CK, Tsai HJ, Liu CC, Lee TH, Hwang PP. Salinity-dependent expression of a Na+, K+, 2Cl- cotransporter in gills of the brackish medaka Oryzias dancena: A molecular correlate for hyposmoregulatory endurance. Comp Biochem Physiol A. 2010;157:7-18
29. Kang CK, Tsai SC, Lin S-T, Lee TH, Hwang PP. Cystic fibrosis transmembrane conductance regulator (CFTR): an apical marker protein of ionocytes for identifying hypoosmoregulation in gills of the euryhaline medaka, Oryzias dancena. Zool Stud. 2012;51:1270-81
30. Kang CK, Yang WK, Lin ST, Liu CC, Lin HM, Chen HH. et al. The acute and regulatory phases of time-course changes in gill mitochondrion-rich cells of seawater-acclimated medaka (Oryzias dancena) when exposed to hypoosmotic environments. Comp Biochem Physiol A. 2013;164:181-91
31. Kang CK, Lee TH. Medaka villin 1-like protein (VILL) is associated with the formation of microvilli induced by decreasing salinities in the absorptive ionocytes. Frontiers in Zoology. 2014;11:2
32. Takehana Y, Naruse K, Sakaizumi M. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol Phylogen Evol. 2005;36:417-28
33. Yang WK, Kang CK, Chen TY, Chang WB, Lee TH. Salinity-dependent expression of the branchial Na+/K+/2Cl- cotransporter and Na+/K+-ATPase in the sailfin molly correlates with hypoosmoregulatory endurance. J Comp Physiol B. 2011;181:953-64
34. Yang WK, Hseu JR, Tang CH, Chung MJ, Wu SM, Lee TH. Na+/K+-ATPase expression in gills of the euryhaline sailfin molly, Poecilia latipinna, is altered in response to salinity challenge. J Exp Mar Biol Ecol. 2009;375:41-50
35. Tang CH, Wu WY, Tsai SC, Yoshinaga T, Lee TH. Elevated Na+/K+-ATPase responses and its potential role in triggering ion reabsorption in kidneys for homeostasis of marine euryhaline milkfish (Chanos chanos) when acclimated to hypotonic fresh water. J Comp Physiol B. 2010;180:813-24
36. Teranishi K, Kaneko T. Spatial, cellular, and intracellular localization of Na+/K+-ATPase in the sterically disposed renal tubules of Japanese eel. J Histochem Cytochem. 2010;58:707-19
37. Tang CH, Leu MY, Yang WK, Tsai SC. Exploration of the mechanisms of protein quality control and osmoregulation in gills of Chromis viridis in response to reduced salinity. Fish Physiol Biochem. 2014;40:1533-46
38. Tresguerres M, Parks SK, Katoh F, Goss GG. Microtubule-dependent relocation of branchial V-H+-ATPase to the basolateral membrane in the Pacific spiny dogfish (Squalus acanthias): a role in base secretion. J Exp Biol. 2006;209:599-609
39. Lin CH, Tsai RS, Lee TH. Expression and distribution of Na, K-ATPase in gill and kidney of the spotted green pufferfish, Tetraodon nigroviridis, in response to salinity challenge. Comp Biochem Physiol A. 2004;138:287-95
40. Kato A, Muro T, Kimura Y, Li S, Islam Z, Ogoshi M. et al. Differential expression of Na+-Cl- cotransporter and Na+-K+-Cl- cotransporter 2 in the distal nephrons of euryhaline and seawater pufferfishes. Am J Physiol Regul Integr Comp Physiol. 2011;300:R284-R97
41. Lubarski I, Pihakaski-Maunsbach K, Karlish SJD, Maunsbach AB, Garty H. Interaction with the Na,K-ATPase and tissue distribution of FXYD5 (related to ion channel). J Biol Chem. 2005;280:37717-24
42. Nebel C, Romestand B, Nègre-Sadargues G, Grousset E, Aujoulat F, Bacal J. et al. Differential freshwater adaptation in juvenile sea-bass Dicentrarchus labrax: involvement of gills and urinary system. J Exp Biol. 2005;208:3859-71
43. Deane EE, Woo NYS. Differential gene expression associated with euryhalinity in sea bream (Sparus sarba). Am J Physiol Regul Integr Comp Physiol. 2004;287:R1054-R63
44. Herrera M, Vargas-Chacoff L, Hachero I, Ruíz-Jarabo I, Rodiles A, Navas JI. et al. Osmoregulatory changes in wedge sole (Dicologoglossa cuneata Moreau, 1881) after acclimation to different environmental salinities. Aquac Res. 2009;40:762-71
45. Fuentes J, Soengas JL, Rey P, Rebolledo E. Progressive transfer to seawater enhances intestinal and branchial Na+-K+-ATPase activity in non-anadromous rainbow trout. Aquac Int. 1997;5:217-27
46. Arjona FJ, Vargas-Chacoff L, Ruiz-Jarabo I, Martín del Río MP, Mancera JM. Osmoregulatory response of Senegalese sole (Solea senegalensis) to changes in environmental salinity. Comp Biochem Physiol A. 2007;148:413-21
47. Freire CA, Amado EM, Souza LR, Veiga MPT, Vitule JRS, Souza MM. et al. Muscle water control in crustaceans and fishes as a function of habitat, osmoregulatory capacity, and degree of euryhalinity. Comp Biochem Physiol A. 2008;149:435-46
48. Li C, Grosdidier A, Crambert G, Horisberger J-D, Michielin O, Geering K. Structural and functional interaction sites between Na,K-ATPase and FXYD proteins. J Biol Chem. 2004;279:38895-902
49. Cornelius F, Mahmmoud YA, Christensen HRZ. Modulation of Na,K-ATPase by associated small transmembrane regulatory proteins and by lipids. J Bioenerg Biomembr. 2001;33:415-23
50. Bibert S, Liu C-C, Figtree GA, Garcia A, Hamilton EJ, Marassi FM. et al. FXYD proteins reverse inhibition of the Na+-K+ pump mediated by glutathionylation of its β1 subunit. J Biol Chem. 2011;286:18562-72
51. Crambert G, Füzesi M, Garty H, Karlish S, Geering K. Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc Natl Acad Sci USA. 2002;99:11476-81
52. Béguin P, Crambert G, Guennoun S, Garty H, Horisberger JD, Geering K. CHIF, a member of the FXYD protein family, is a regulator of Na,K-ATPase distinct from the γ-subunit. EMBO J. 2001;20:3993-4002
53. Béguin P, Crambert G, Monnet-Tschudi F, Uldry M, Horisberger JD, Garty H. et al. FXYD7 is a brain-specific regulator of Na,K-ATPase α1-β isozymes. EMBO J. 2002;21:3264-73
54. Crambert G, Li C, Claeys D, Geering K. FXYD3 (Mat-8), a New Regulator of Na,K-ATPase. Mol Biol Cell. 2005;16:2363-71
55. Garty H, Lindzen M, Scanzano R, Aizman R, Füzesi M, Goldshleger R. et al. A functional interaction between CHIF and Na-K-ATPase: implication for regulation by FXYD proteins. Am J Physiol Renal Physiol. 2002;283:F607-F15
56. Nasevicius A, Ekker SC. Effective targeted gene `knockdown' in zebrafish. Nat Genet. 2000;26:216-20
57. Varsamos S, Nebel C, Charmantier G. Ontogeny of osmoregulation in postembryonic fish: a review. Comp Biochem Physiol A. 2005;141:401-29
Author contact
![]() Corresponding author: THL, Department of Life Sciences, National Chung Hsing University, 145 Xingda Road, Taichung 402, Taiwan. Tel: +886-4-2285-6141; fax: +886-4-2287-4740; E-mail: thleenchu.edu.tw.
Corresponding author: THL, Department of Life Sciences, National Chung Hsing University, 145 Xingda Road, Taichung 402, Taiwan. Tel: +886-4-2285-6141; fax: +886-4-2287-4740; E-mail: thleenchu.edu.tw.

 Global reach, higher impact
Global reach, higher impact