Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(6):757-767. doi:10.7150/ijbs.14333 This issue Cite
Research Paper
A Novel Protein Elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 Induces Systemic Resistance in Tobacco
The State Key Laboratory for Biology of Plant Disease and Insect Pests, Institute of Plant protection, Chinese Academy of Agricultural sciences, No. 12 Zhongguancun South Street, Beijing 100081, China
Received 2015-11-6; Accepted 2016-2-2; Published 2016-4-28
Abstract

Here we reported a novel protein elicitor from Bacillus amyloliquefaciens NC6 induced systemic resistance (ISR) in tobacco. The purification was executed by ion-exchange chromatography, native-page extraction and HPLC, and the amino acid sequence was identified by mass spectrometry. This recombinant elicitor protein, expressed in Escherichia coli by an E1 expression vector, had good thermal stability, and the elicitor caused a clearly defined hypersensitive response (HR) necrosis in tobacco leaves. It could also trigger early defence events, including generation of reactive oxygen species (H2O2 and O2-) and phenolic-compound accumulation. Quantitative real-time PCR (Q-RT-PCR) results indicated that several plant defence genes, including the salicylic acid (SA)-responsive PR1a, PR1b, PR5, and phenylalanine ammonia lyase (PAL), as well as the jasmonic acid (JA)-responsive PDF1.2 and CORONATINE INSENSITIVE 1 (COI1), were all up-regulated. Moreover, infiltration conferred systemic resistance against a broad spectrum of pathogens, including Tobacco mosaic virus (TMV) and the fungal pathogen Botrytis cinerea.
Keywords: protein elicitor, induced systemic resistance, ROS, Hypersensitive response
Introduction
Pathogen attacks are responsible for losses in agricultural yield. Crops, growing naturally in soil, are surrounded by various microorganisms that can be beneficial or pathogenic. Pathogenic microorganisms can infect plants and cause severe disease. However, beneficial bacteria, such as the plant growth-promoting rhizobacteria (PGPR), are among the various groups of plant-associated microorganisms that can reduce the activity of pathogenic microorganisms by microbial antagonism through competition for nutrients, production of antibiotics and secretion of lytic enzymes (1,2,3). Furthermore, PGPR can reduce the severity of disease or pathogen multiplication by indirectly eliciting the plant defence system (4), which has evolved from a long-term interaction with microorganisms (5, 6). This resistance response phenomenon is called the induced systemic resistance (ISR), which can be locally induced by beneficial microbes, and enhance broad spectrum systemic resistance to plant pathogens (7).
The spectrum of diseases to which PGPR-elicited ISR confers enhanced resistance overlaps partly with that of pathogen-induced systemic acquired resistance (SAR), which is induced by pathogen-associated molecular patterns (PAMPs) (8). Both ISR and SAR represent a state of enhanced basal resistance in the plant that depends on the signalling molecules jasmonic acid (JA) and salicylic acid (SA), respectively. These two molecules play important roles in the regulation of signalling networks in induced defence responses (9, 10). Normally, rhizobacteria-mediated ISR is independent of SA but requires JA and ethylene signalling in the plant (11). ISR is not commonly associated with an accumulation of pathogenesis-related (PR) proteins. However, in other cases, activation of an SA-dependent pathway is a feature of ISR-inducing PGPR, and ISR by Bacillus spp. and Pseudomonas protegens lead to accumulation of the defence gene PR1 in plants (12, 13, 14). Pathogens are differentially sensitive to the resistances activated by each of these signalling pathways (15).
PGPR and avirulent pathogens induce resistance responses by releasing elicitors to plants. These elicitors can be biotic or abiotic; include proteins, (poly)peptides, lipids, and oligosaccharides (16,17,18,19,20,21); and are molecules that are capable of mimicking the perception of a pathogen by a plant, thereby triggering a sophisticated defence response in plants (22, 23). The elicitor proteins such as, harpins and elicitins are usually derived from pathogenic organisms like gram-negative bacteria (17) and oomycetes (24), only a few of them are isolated from PGPR.
Elicitors derived from pathogenic bacteria and PGPR can induce a series of responses in plants. The hypersensitive reaction (HR) is a typical characteristic caused by elicitors in plant leaves, which is a part of the plant innate immunity and aims to limit the invading pathogens to the infected area by rapid and localized programmed cell death (PCD) (25, 26). This response causes the induction of signalling cascades and changes. Another responses including an oxidative burst concomitant with the generation of reactive oxygen species (ROS) and nitrogen species (NO) in tobacco leaves and cell suspensions. The production of NO induced by elicitors is partly regulated through an ROS-dependent pathway involving the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase. In turn, NO can down-regulate the level of H2O2 (27, 28). Activation of defence genes and the production of antimicrobial secondary metabolites, such as phytoalexins, papillae formation, callose and phenolic compounds, act as plant barriers to pathogens and ultimately, lead to resistance to the pathogen (29, 30, 31).
Many of the ISR-inducing microbes identified so far are Gram-negative bacteria and more particularly species belonging to the Pseudomonas. However, the number of Bacillus spp., reported as ISR inducers has grown rapidly over the last decade and include B. amyloliquefaciens, Bacillus subtilis, Bacillus pasteurii, Bacillus cereus, Bacillus pumilus, Bacillus mycoides, and Bacillus sphaericus. These species can elicit significant reductions in disease severity for a broad range of pathogens in a diversity of hosts. Dimethyl disulfide (DMDS), produced by a B. cereus strain, has ISR-eliciting activity and leads to resistance to plant fungal diseases (32). Volatile chemicals produced by the PGPR strains B. subtilis GB03 and B. amyloliquefaciens IN937a are reported to trigger plant defence responses (33). Fengycins and surfactins derived from B. subtilis can also induce plant defence responses (34).
We report the purification and characterization of novel protein elicitor PeBA1 from the B. amyloliquefaciens, a well-studied PGPR strain for biocontrol in crop cultivation. We performed a purification process that consisted of ion-exchange chromatography, native-page extraction, and high-performance liquid chromatography (HPLC). The amino acid sequence was identified by mass spectrometry, and the protein was then heterologously expressed in Escherichia coli. The recombinant protein induced early signalling events of the plant defence responses in tobacco and triggered systemic resistance against infection by Tobacco mosaic virus (TMV) and Botrytis cinerea.
Materials and Methods
Plant, bacteria, and pathogens culture conditions
Nicotiana benthamiana and Nicotiana tabacum were grown at 24-26 °C, with a 12-h light/dark in a phytotron. Tobacco cells were cultured at 25 °C in Murashige and Skoog medium (4.2g/L Murashige and Skoog, 100 mg/ml inositol, 0.2% KH2PO4, 0.2 mg/ml 2, 4-dichlorophenoxyacetic acid, and 3% sucrose, and 1mg/L VB1, pH 5.2) in a shaker at 150 rpm in the dark. B. amyloliquefaciens NC6 strain was grown on LB medium at 37 °C. B. cinerea was maintained on potato dextrose agar medium (PDA) at 25 °C in the dark. TMV was cultivated in N. tabacum for bioassays.
Protein purification
B. amyloliquefaciens NC6 was cultivated in 1 L of LB medium and shaken at 200 rpm for 48 h at 37 ºC. The supernatant was collected, precipitated with ammonium sulfate at 30% saturation overnight at room temperature, re-suspended in 50 ml buffer A (50 mM Tris-HCl, pH 8.3), and dialysed at 4 ºC for 48 h. Total protein was filtered with a 0.22-µm membrane (Millipore, Corp., Billerica, MA, USA), and further purification with the Äkta Explorer 10 protein purification System (GE Healthcare, Piscataway, NJ, USA) used an ion-exchange chromatography column (HP Q HiTrapTM 5 ml, GE Healthcare, Uppsala, Sweden), loading buffer was buffer A, and elution buffer was buffer B (50 mM Tris-HCl, 1 M NaCl). A linear gradient was eluted, and all fractions were collected. The samples were desalted using a 10-kDa ultrafiltration device and were tested for induced tobacco leaf HR activity. We concentrated the active fraction and then used native polyacrylamide gel electrophoresis (native-PAGE) for further purification. The single distinct protein band was retrieved for the HR test, further purification via HPLC, and to monitor elicitor activity.
PeBA1 mass spectrometry analysis, gene cloning and expression
The protein sample isolated on a sodium dodecyl sulphate (SDS)-PAGE gel was identified by mass spectrometry (MS) analysis (Beijing Protein Innovation, Beijing, China). The tandem MS (MS-MS) data were automatically analysed using the Mascot search engine (Matrix Science, London, United Kingdom).
The genomic DNA was isolated from B. amyloliquefaciens NC6 using an E.Z.N.A. bacterial DNA kit (Omega Bio-Teach, Norcross, GA, USA). According to the results from the mass spectrographic analysis, we designed a pair of primers (Table 1) to clone the gene from B. amyloliquefaciens NC6. This gene was then cloned into the E1 plasmid (TransGen Biotech, Beijing, China), which has 18aa in upstream (MRGSHHHHHHGMASELAL), and transformed into E. coli BL21(DE3) cells (TransGen Biotech, Beijing, China).
To express the recombinant PeBA1, Transformed BL21 cultures grown overnight and then transferred single colony into LB medium, growing at 37 ºC. Then, when the OD600 was 0.6, 100 μM isopropyl β-D-1-thiogalactopyranoside (IPTG, Sigma, St. Louis, MO, USA) was added to induce expression, and the temperature was changed to 16 ºC with shaking at 200 rpm overnight.
Bacteria was harvested by centrifugation, re-suspended in buffer C (50 mM Tris-HCl, 200mM NaCl, pH 8.3) and disrupted three times with an ultrasonic disruptor. The supernatant containing the recombinant elicitor was collected through centrifugation at 12,000 g for 30 min. Further purification of PeBA1 was executed by affinity chromatography with a His-Trap HP column (GE Healthcare, Waukesha, WI, USA), loading buffer is buffer C, eluted by buffer D (50 mM Tris-HCl, 200mM NaCl, 500 mM imidazole, pH 8.3) directly, followed by desalting with buffer E (50 mM Tris-HCl, pH 8.3) in a HiTrap desalting column (GE Healthcare, Waukesha, WI, USA). The molecular mass was monitored by 12% SDS-PAGE, and a protein marker (Thermo Scientific, Rockford, IL, USA) was used to estimate the molecular masses of the purified recombinant elicitor.
Characterization of PeBA1
In order to assay the effects of recombinant PeBA1 for the induction of HR in tobacco, 20 µM recombinant PeBA1 was infiltrated into the leaves using a needless syringe. HR symptoms were examined after 24 h. Different concentrations of recombinant PeBA1 (20, 10, 5, 1, and 0.5 μM) in a 100-μl volume were infiltrated into tobacco leaves to determine the minimum concentration of recombinant PeBA1 needed for HR activity, with buffer alone used as a control.
To further investigate cell death, trypan blue staining was performed by boiling leaf tissues in a mixture of phenol, lactic acid, glycerol and distilled water containing 1 mg/ml trypan blue (1:1:1:1) for 1 min. The samples were then soaked in 2.5 mg/ml chloral hydrate overnight (35). HSR203J, the HR marker gene of tobacco, was amplified with RT-PCR (36) using Actin gene as a positive control. Primers were designed for HSR203J and Actin (Table 1).
To test the effects of temperature and pH on our protein, we treated recombinant PeBA1 at different temperatures (4, 25, 50, 80, and 100 ℃) for 15 min and different pH values (4, 6, 8, 10, and 12) overnight. The HR responses for infiltrated tobacco leaves were observed at 24 h.
Primers used for PCR and RT-PCR in this study
| Gene name | Forward primer(5' to 3' ) | Reverse primer(5' to 3') |
|---|---|---|
| PeBA1 HSR203J | ATGGCTAACCAGAAGAAGAAAAC TGTACTACACTGTCTACACGC | TTATCCGTTTACGATGGTGC GATAAAAGCTATGTCCCACTCC |
| Actin | AGCAACTGGGATGACATGGA | GCGGTGATTTCCTTGCTCAT |
Primers used for RT-qPCR in this study
| Gene name | Forward primer(5' to 3' ) | Reverse primer(5' to 3') |
|---|---|---|
| PR1a | GATGCCCATAACACAGCTCG | CGAGGTTACAATCTGCAGCC |
| PR1b | GACAAGTTGGTGTTGGTCCC | GGCTAGGTTTTCGCCGTAAG |
| PR5 | AGTGGCCGAGGTAATTGTGA | CATTGGTCGGGCTGAATTCC |
| PAL | ATTGCTGGTTTGCTCACTGG | TCCTTAGGCTGCAACTCGAA |
| NPR1 | GATACACGGTGCTGCATGTT | AAGCCTAGTGAGCCTCTTGG |
| PDF1.2 | ATCTGTCTGGGGAAATGGCA | CATGGTCCCTTGAAACGGTG |
| COI1 | AACTGGTCGGGATCTCTTGG | TAGGCAAGTATATGGGCGGG |
| Actin | CGAGGGTTATGCTTTGCCTC | AGCAAGCTCCTCCTTCATGT |
The induction of oxidative burst
The ROS burst, such as the generation of H2O2 and O2-, was assayed with 3,3'-diaminobenzidine (DAB) and Nitroblue Tetrazolium (NBT) (37), respectively. Leaves of six-week-old tobacco plants were infiltrated with recombinant PeBA1 through stomata. After four hours, the leaves were cut and then vacuum-infiltrated with 1 mg/ml DAB (pH 3.8) or 0.5 mg/ml NBT for 1 h. The treated tissues were incubated for more than 12 h in ethanol to eliminate chlorophyll, observed for DAB and NBT deposits, and photographed.
H2O2 accumulation in tobacco cell was quantified by chemiluminescence using luminol and a GloMax-96 luminometer (Promega, Madison, WI, USA) (38). Seven-day-old cells were harvested and re-suspended (1 g cell for 10 ml buffer) in buffer (10 mM HEPES, 175 mM mannitol, 0.5 mM K2SO4 and 0.5 mM CaCl2, pH 5.75). After incubation for 1 h at 26 ºC on a rotary shaker (160 rpm), a 250 μl aliquot of the tobacco cell suspension was added to 300 μl assay buffer containing 50 μl of 0.3 mM luminol. One micromolar recombinant PeBA1 was added to the assay buffer to elicit a response, and measurements were taken at the indicated times.
Detection of phenolic-compound accumulation
To visualize phenolic-compound accumulation in the tobacco suspension cells as described (39), approximately 300 μl of tobacco cell suspension was incubated with 1 µM recombinant PeBA1 (using buffer as a negative control) on a shaker in the dark at 26 °C for 108 h. The cell was then examined with a confocal microscope (LSM 510; Zeiss, Oberkochen, Germany).
Bioassay for PeBA1-induced disease resistance in tobacco
Six-week-old N. tabacum leaves were treated with 20 μM recombinant PeBA1, using buffer as a control. Three days later, systemic leaves were mechanically inoculated with TMV. Inoculated plants were maintained at 25 °C under a 12-h light/dark regime and inspected daily for disease symptoms. The disease severity was recorded by measuring lesion diameters and by counting the number of TMV lesions at 3 days post-inoculation (dpi). For the diameters of the lesions, 10 random lesions were measured for each plant leaf.
Inoculation of B. cinerea was performed using mycelia discs. Systemic leaves were detached in 2 days after recombinant PeBA1 treatment. Mycelia discs were transferred to detached N. benthamiana, and inoculated leaves were then placed in Petri dishes containing 1.5% agar medium and kept in a high-humidity chamber. Disease severity was recorded by measuring diameter sizes.
Gene expression analysis
To clarify the mechanisms of the defence responses induced by recombinant PeBA1 in tobacco plants, a certain concentration (20 μM) of recombinant PeBA1 (using buffer as a control) was infiltrated into six-week-old N. tabacum leaves. Systemic leaves were excised at the indicated times. Total RNA from tobacco was isolated using the RNAprep pure Plant Kit (Tiangen Biotech, Beijing, China). First-strand cDNA was synthesized using the FastQuant RT Kit (Tiangen Biotech, Beijing, China) with 1.5 μg of total RNA as the template. Quantitative real time PCR (Q-RT-PCR) was performed on an IQ-5 real-time system (Bio-Rad Laboratories, Hercules, California, USA) using SuperReal PreMix Plus (Tiangen Biotech, Beijing, China). Actin was used as an internal reference to standardize the RNA samples for evaluating relative expression levels. Relative expression was calculated using the 2-ΔΔCT method (40). All primers are described in table 1.
Protein assay
All protein concentrations were measured using a Bradford assay.
Statistical analysis
All experiments were repeated independently three times, and the data are presented as the means and standard deviations. Significant differences between the treatments and the controls were determined using the Student's t-test and SAS software (SAS Institute Inc., Cary, NC, USA).
Results
Purification, characterization and identification of PeBA1
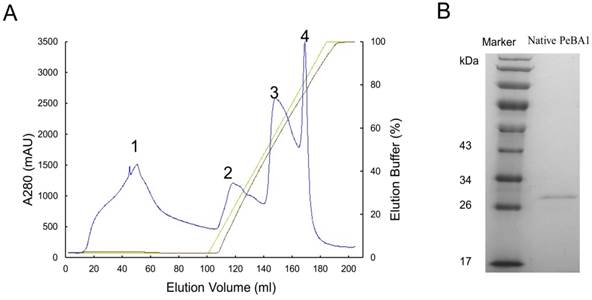
Crude protein from B. amyloliquefaciens NC6 was dialysed, and anion exchange chromatography was performed. Proteins have different affinity for chromatographic column, a linear gradient eluting with buffer B can elute different protein gradually according affinity from low to high. We collected four protein peaks (Fig. 1A), first peak is filtrate proteins which had no affinity to column, other peaks were eluting peaks, and every peak was desalted and infiltrated into tobacco leaves for an HR test (data not shown). Peak 2 had HR activity. This activity peak was isolated using native-PAGE and HPLC for further purification, and we found just one clear protein band between 26 kDa and 34 kDa (Fig. 1B). This band was retrieved and had HR activity. HPLC showed only one peak, and we ran SDS-PAGE again for mass spectrometry analysis.
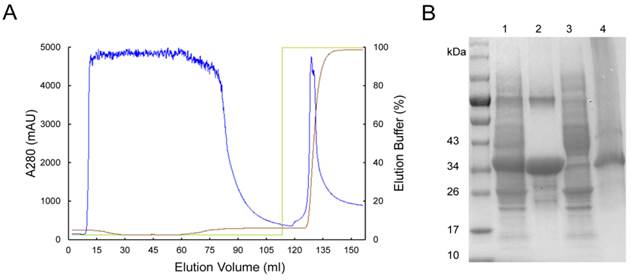
The single protein band was excised from the SDS-PAGE gel for identification by liquid chromatography-mass spectrometry analysis. The results were searched by Mascot, and we obtained the best-matching protein (GenBank: KT362141). This protein has 285 amino acid residues, contains a SDR domain, and belongs to the short-chain dehydrogenase family. The full-length PeBA1 (858bp) was cloned. Transformed into E1 vector. And expressed in E. coli BL21(DE3). Recombinant protein elicitor was purified by affinity chromatography (Fig. 2A) and desalted. The recombinant protein exhibited a single band with an apparent molecular mass of approximately 34 kDa on an SDS-PAGE gel (Fig. 2B), which was in agreement with the calculated size of the fusion protein.
PeBA1 induced HR in non-host plants
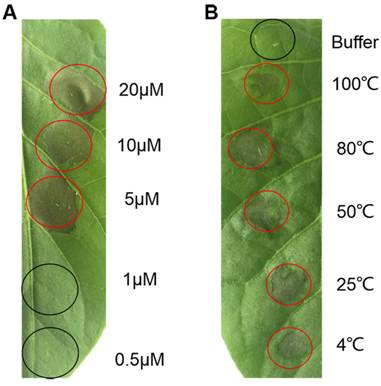
Recombinant PeBA1 was infiltrated into leaves of tobacco through the stoma. There were clearly defined HR necrotic areas at the infiltration site at 24 h post-infiltration (hpi) (Fig. 3A). The corresponding protein buffer had no effect on the infiltrated leaves. Serial dilutions of recombinant PeBA1 were infiltrated into leaves for HR testing, and the results indicated that the minimum concentration of the elicitor that induced HR was 5 μM (Fig. 4A).
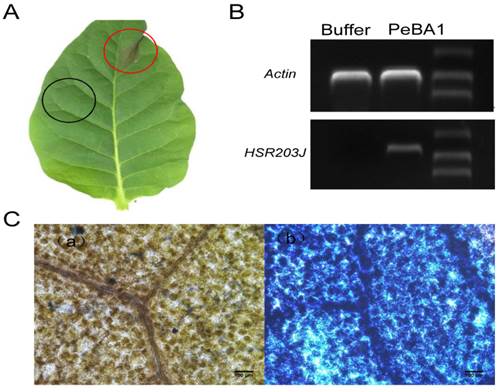
Recombinant PeBA1 also induced apparent expression of the HR marker gene HSR203J in treated tobacco leaves (Fig. 3B), HR is also a form of cell death, which can be monitored by trypan blue staining on the leaves. Thus, dead cells located at the site of HR were stained blue (Fig. 3C).
We found that recombinant PeBA1 was stable at 4, 25, 50, 80 and 100 ℃ for 15 min and retained its activity to induce HR in leaves (Fig. 4B). In addition, the protein elicitor exhibited HR activity over a broad spectrum of pH ranges. After incubation at 4 °C overnight in different pH solutions (pH 4, 6, 8, and 12), all infiltrated areas showed obvious HR activity in tobacco leaves.
Purification of PeBA1 from Bacillus amyloliquefaciens NC6. (A) Crude protein from the ammonium sulfate precipitation was purified using the Akta Explorer 10 protein purification system. Using an ion-exchange chromatography column, four peaks (1, 2, 3, and 4) were collected. (B) Peak 2 was further purified by native-PAGE and HPLC. PeBA1 was resolved on an SDS-PAGE gel with a protein molecular mass between 26 and 34 kDa.
Purification of recombinant PeBA1. (A) Total protein expressed in E. coli was purified by the Akta Explorer 10 protein purification system using a His-Trap HP column. Recombinant PeBA1 was eluted with high-concentration imidazole. (B) Expression and purification of recombinant PeBA1. 1: total protein, 2: soluble recombinant PeBA1, 3: filtrate, 4: disrupted cell pellet.
PeBA1-induced HR in plant leaves. (A) HR in tobacco leaves was observed at 24 hours post-infiltration with PeBA1. Red circles were treated with 30 μl PeBA1 (20μM) and showed obvious HR, while buffer (Tris-HCl, pH8.3) as a control (black circles) did not induce HR. (B) RT-PCR was executed at 12 h after treatment with PeBA1 and buffer in tobacco leaves and HSR203J gene expression was induced by PeBA1. (C) PeBA1-induced cell death in infiltrated areas was stained blue (b), buffer treatment areas could not be stained by dye (a). Scale bar = 100 μm.
The minimum concentrations of PeBA1 for HR activity and thermo-stability. (A) Serial dilutions of PeBA1 (20, 10, 5, 1, and 0.5 μM /L) were infiltrated into tobacco leaves, and HR was observed after 24 hours. (B) PeBA1 (50 μl of 20 μM/L) was treated at different temperatures (4, 25, 50, 80, and 100℃) for 15 min, infiltrated into leaves through stoma (using buffer as a control), and photographed at 24 hpi. Infiltrated areas are labelled with circles, and red circles showed typical HR.
PeBA1 caused ROS accumulation
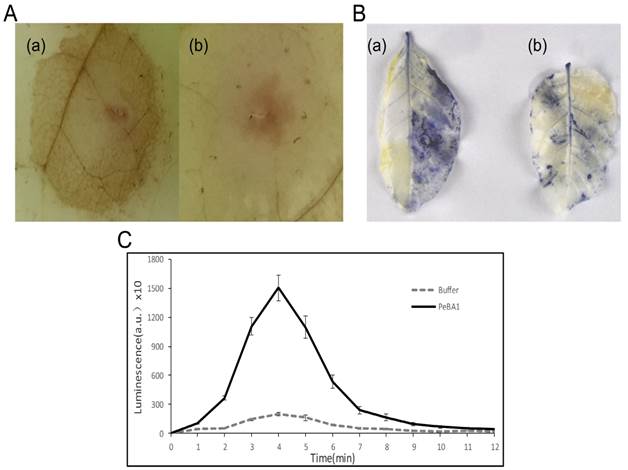
ROS burst is one of the earliest significant events before the HR (41) and has a crucial role in plant systemic resistance (42). To further examine the recombinant PeBA1-activated HR biochemical responses, we analysed the accumulation of ROS generated by the plasma membrane-localized NADPH oxidase complex (43). H2O2 and superoxide anion (O2-) production levels were assayed using DAB and NBT, respectively. Significant brown DAB-stained precipitates were easily and clearly observed in the recombinant PeBA1-treated area (Fig. 5A), and a blue precipitate indicated an abundance of O2- production (Fig. 5B) in tobacco leaves after infiltration with PeBA1 at 4 hpi.
H2O2 production induced by recombinant PeBA1 was measured with a tobacco cell suspension using chemiluminescence. H2O2 production peaked 4 min after recombinant PeBA1 addition and then gradually decreased to a level slightly above buffer-treated levels at 12 min (Fig. 5C).
PeBA1 induced phenolic-compound accumulation
To determine whether recombinant PeBA1 triggered the plant defence response through the accumulation of secondary metabolites that act as plant barriers, we examined phenolic-compound accumulation in tobacco after treatment with recombinant PeBA1.
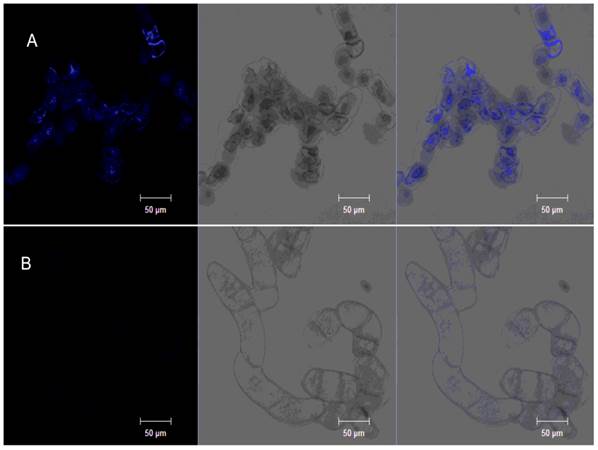
Phenolic compounds, which are secondary metabolism molecular components, contribute to the control of plant diseases. Phenolic compound accumulation in a tobacco cell suspension could be visualized with ultraviolet (UV) excitation (44). Phenolic compounds accumulated in the tobacco cell suspension were observed as blue auto-fluorescence (Fig. 6A).
Induction of ROS in tobacco leaves by PeBA1. (A) H2O2 accumulation in leaves was stained with DAB at 4 h post-inoculation (hpi). (a) PeBA1-infiltrated areas were stained brown compared with (b) buffer treatment as a control. (B) NBT revealed the production of O2-, and staining was performed on leaves at 4 hpi. O2- accumulation appeared in PeBA1-treated leaves (a), but not in leaves treated with buffer (b). (C) Kinetics of ROS production following elicitation in tobacco cell culture. ROS production was measured by chemiluminescence. PeBA1 rapidly induced ROS formation, with the peak occurring at 4 min. The kinetics are representative of three independent experiments. Error bars represent ± SD of the mean. Values are expressed in arbitrary units (a.u.).
PeBA1-induced phenolic-compound accumulation. Tobacco cell suspension was incubated with PeBA1 or buffer for 108 h. Obvious intense blue fluorescence appeared in the PeBA1-treated cell culture (A) compared with buffer treatment (B). Samples were photographed by confocal microscopy under different lights: fluorescence (left), bright (middle) and overlay (right). Three independent biological replicates were used for the suspension-cultured tobacco cells.
PeBA1 induced plant systemic resistance against Botrytis cinerea and TMV
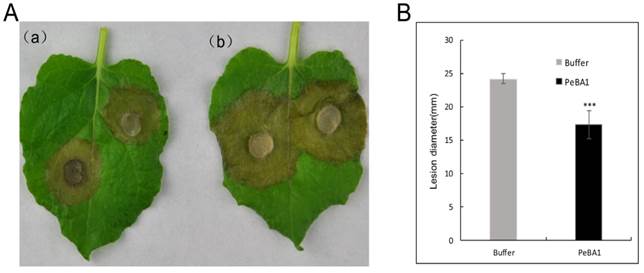
We tested the capacity of recombinant PeBA1 to induce systemic disease resistance against the fungal pathogens B. cinerea and TMV. N. benthamiana and N. tabacum leaves were infiltrated with recombinant PeBA1. The recombinant PeBA1 treatment significantly inhibited the expansion of lesions in the B. cinerea-inoculated leaves (Fig. 7) at 96 hours post infection.
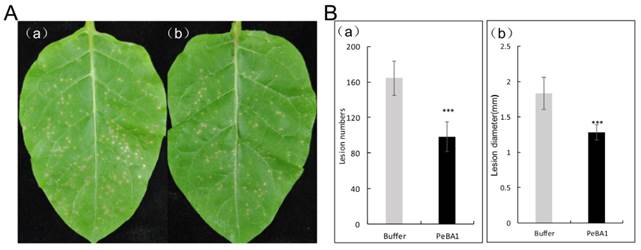
To determine if recombinant PeBA1 triggered resistance to TMV, N. benthamiana leaves were inoculated with TMV. By 3 dpi, plant leaves, which were treated with recombinant PeBA1, exhibited obvious disease resistance. Both the number and diameters of TMV lesions in systemic leaves of the recombinant PeBA1-treated plants were obviously fewer and smaller, respectively, than those of the control plants (Fig. 8).
PR genes are up-regulated by PeBA1
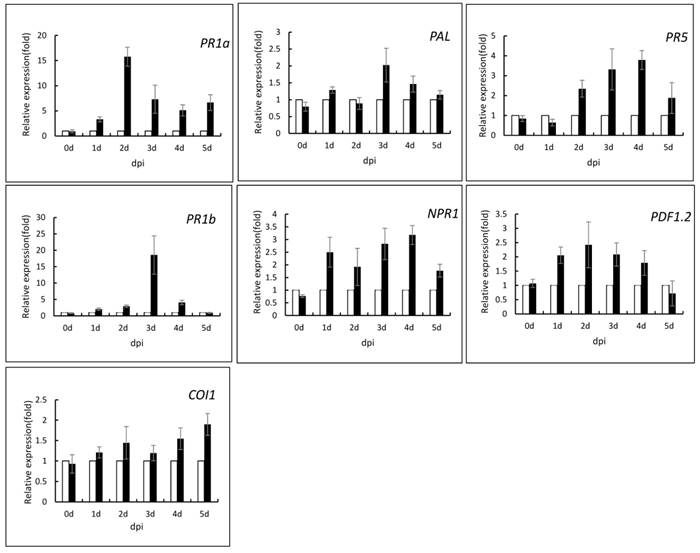
To investigate the underlying mechanism of recombinant PeBA1-induced plant resistance, we infiltrated leaves of 6-week-old N. tabacum with recombinant PeBA1 proteins and evaluated the expression levels of several plant defence-related genes. The pathogenesis-related proteins (PR1a, PR1b and PR5) and phenylalanine ammonia lyase (PAL), which are all marker genes of the SA-dependent defence pathway, were significantly up-regulated in response to recombinant PeBA1 (Fig. 9). The expression levels of the PR1a gene were significantly up-regulated at 2 dpi, and the maximum level of the gene increased by 15-fold; the level then decreased but was also up-regulated compared with the control. PR1b was significantly up-regulated by approximately 20-fold at 3 dpi. Expression of the PR5 gene continuously increased from 2 to 4 dpi. Meanwhile, recombinant PeBA1 treatment triggered the expression of CORONATINE INSENSITIVE 1 (COI1) and up-regulation of the plant defensin gene 1.2 (PDF1.2) (Fig. 9), which are JA-responsive. The expression level of PDF1.2 was increased 2-fold after recombinant PeBA1 treatment and was continuously up-regulated for 4 days; COI1 was also up-regulated. Non-expressor of pathogenesis related 1 (NPR1) was up-regulated at 1 to 4 days (Fig. 9). This gene product not only plays an essential role in SA-mediated systemic acquired resistance but also has an important role in rhizobacterium-triggered ISR (45).
Discussion
B. amyloliquefaciens is a kind of plant root-colonizing PGPR. A series of studies has elucidated several aspects of this rhizobacterium in plant disease biocontrol. Here, we reported that a novel protein elicitor, PeBA1, obtained and purified from the B. amyloliquefaciens NC6 strain, Recombinant proteins of PeBA1 obtained from E. coli can induce cell death, transcription of defence genes, ROS production, HR necrosis, and accumulation of phenolic compounds, displaying features of ISR in tobacco.
PeBA1 has 285 amino acid residues, contains a SDR domain, and belongs to the short-chain dehydrogenase family. However, a signal peptide was not found, which indicated that this elicitor is secreted by another secretory pathway. Tjalsma and co-workers discovered that approximately 26% of the extracellular proteome lacks typical signal peptides and could escape from the cytoplasm by cell lysis or via the flagellar export machinery (46). Dehydrogenases, including pyruvate dehydrogenase E1 alpha subunit pdhA/B, leucine dehydrogenase bcd and malate dehydrogenase mdh, are B. amyloliquefaciens secretion proteins that lack a signal peptide (47).
PeBA1-induced resistance in tobacco against Botrytis cinerea. (A) Representative phenotypes of disease caused by B. cinerea in PeBA1- and buffer-induced tobacco leaves. The sizes of the lesions caused by B. cinerea in PeBA1-induced leaves (a) were smaller than in buffer-induced leaves (b) at 96 h post-infiltration. B. Lesion sizes caused by B. cinerea were measured in leaves from PeBA1- or buffer-induced plants. Data presented in (B) are the means ± SD from three independent experiments. The statistical analyses were performed using Student's t test, and asterisks indicate significant differences between PeBA1 and buffer treatment (***, P < 0.001)
PeBA1-induced systemic resistance in tobacco against TMV. Three days after infiltration with PeBA1 or buffer (as a control) on leaves of 6-week-old tobacco plants, systemic leaves were inoculated with TMV. Photographs were taken at 3 days post-inoculation (dpi). (A) Representative phenotypes of disease symptoms caused by TMV infection in tobacco leaves. (A) The numbers and diameters of TMV lesions in systemic leaves of PeBA1-induced plants (b) were significantly fewer and smaller, respectively, than those in control plants (a). (B) Systemic resistance conferred by PeBA1. The numbers of local lesions caused by TMV infection were counted, and lesion sizes were measured at 3 dpi. Values are the means ± SD from three independent experiments. ***P < 0.001 by Student's t-test.
Expression analysis of defence-related genes in tobacco after PeBA1 treatment using buffer as a control. The samples were harvested from systemic leaves at the indicated times, and Q-RT-PCR was performed to show the relative expression levels of the PR-1a, PR1b PR-5, PAL, NPR1, PDF1.2, and COI1 genes. The samples were normalized against Actin, and expression levels are represented as fold changes in relation to the control.
HR is part of the plant innate immunity (30), and genetic approaches have elucidated functions of HR in the initiation of resistance genes and association with death-associated signals, including calcium, ROS, NO, SA, and sphingolipids (48). Furthermore, the HSR203J gene, which is considered a marker of HR in tobacco (49), was prominently expressed in tobacco leaves at 12 h post-PeBA1 treatment (Fig. 3B). The area infiltrated with the PeBA1 treatment showed an obvious blue colour after Trypan blue staining (Fig. 3C) (50). These tests indicate that PeBA1 can induce necrosis in several nonhosts with symptoms of HR.
One of the earliest events during the HR is the generation of reactive oxygen as an active process to signal downstream cellular processes (51). ROS include H2O2 and O2- (Fig. 5), whose production results mainly from the activity of NADPH oxidases and superoxide dismutases (SOD), respectively. The main generation of ROS in plants is the ROS burst, which follows plant detection of invading pathogens or pathogen-derived elicitors, also known as PAMPs (52). On the one hand, ROS enhanced strengthening of host cell walls via cross-linking of glycoproteins, on the other hand, ROS production acts as a an intercellular or intracellular signalling molecule that interacts with various molecules, including lipids, Ca2+, NO, mitogen-activated protein kinase (MAPK), and wounding-induced protein kinase (WIPK). These signalling molecules may be involved in defence signalling pathways (35, 53). And ROS metabolism in plant could also provide the redox environment for regulating the function of NPR1, a crucial mediator of systemic responses to plant disease (54).
Recombinant PeBA1 induced resistance in tobacco against a broad spectrum of pathogens, including TMV (Fig. 8) and the fungal pathogen Botrytis cinerea (Fig. 7). Beneficial microorganisms induce systemic defence responses that are controlled by a signalling network in which the plant hormones SA, JA, and ethylene (ET) play important roles (29). Some evidence has shown that the SA, JA, and ET pathways crosstalk to adjust the plant defence response depending on different pathogens (55). In this study, we provide evidence that PeBA1 enhances the expression of several SA-responsive genes, including PR1a, PR1b, PR5 and PAL (Fig. 9), and JA/ET-responsive genes, including PDF1.2 and COI1 (Fig. 9). Furthermore, recombinant PeBA1 enhances NPR1 expression (Fig. 9), which is essential for the induction of PR genes through interaction with TGA transcription factors (56, 57, 58). Pieterse and co-worker demonstrate that signal transduction leading to P. fluorescens-mediated ISR requires responsiveness to JA and ET and is dependent on NPR1 (11). These results demonstrate that SA- and JA/ET-dependent pathways may be involved in the PeBA1-induced systemic defence responses in tobacco.
In addition, secondary metabolite accumulation process can act in plant defense against infection by establishing mechanical barriers to pathogen growth. Elicitor can induce phenolic metabolism, and lignin synthesis. Phenolic compounds, including scopoletin, phenolic acids and cell wall bound ferulic acid (59), can confer mechanical strength to the cell wall to protect against infection with pathogenic bacteria or fungi, ferulic acid crosslinking of phenylpropanoid esters leads to the formation of ligninlike polymers, such hydroxycinnamic acids and their derivatives display autofluorescence (60). Phenylalanine ammonia lyase (PAL) is a key enzyme for the synthesis of various phenolic compounds in the phenylpropanoid pathway (39). Correspondingly, Q-RT-PCR showed that PAL was induced by recombinant PeBA1 in tobacco (Fig. 9).
In summary, we report a novel elicitor protein PeBA1 from B. amyloliquefaciens NC6 strain, which can induce a typical HR in tobacco leaves and activate defence-related early events, such as an ROS burst. After further study, we find that PeBA1 enhances tobacco disease resistance to TMV and Botrytis cinerea by triggering defence-related gene up-regulation and accumulation of antimicrobial compounds. These results indicate that our research helps to elucidate the mechanisms of B. amyloliquefaciens NC6-induced systemic resistance in tobacco.
Supplementary Material
Figure S1.
Acknowledgements
This work was supported by the Special Fund for Agro-Scientific Research in the Public Interest (201303021) and international cooperation project (2014DFG32270).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Handelsman J, Stabb EV. Biocontrol of soilborne plant pathogens. Plant Cell. 1996;8:1855-1869
2. Van Loon LC, Bakker PAHM. Signalling in rhizobacteria-plant interactions. In Root ecology, Springer Berlin Heidelberg. 2003:297-330
3. Van Loon LC, Glick BR. Increased plant fitness by rhizobacteria. In Molecular ecotoxicology of plants. Springer Berlin Heidelberg. 2004:177-205
4. Haas D, Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307-319
5. Diez-Navajas AM, Wiedemann-Merdinoglu S, Greif C. et al. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology. 2008;98:776-780
6. Pieterse CMJ, Leon-Reyes A, Does DVD. et al. Networking by small-molecule hormones in plant immunity. IOBC/WPRS Bulletin. 2012;83:77-80
7. Pieterse C M J, Zamioudis C, Berendsen R L. et al. Induced systemic resistance by beneficial microbes. Annual review of phytopathology. 2014;52:347-375
8. Boller T, Felix G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu Rev Plant Biol. 2009;60:379-406
9. Pieterse CMJ, Van Loon LC. Salicylic acid-independent plant defence pathways. Trends Plant Sci. 1999;4:52-58
10. Glazebrook J. Genes controlling expression of defense responses in Arabidopsis - 2001 status. Curr Opin Plant Biol. 2001;4:301-308
11. Pieterse CMJ, van Wees SCM, van Pelt JA. et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571-1580
12. Kloepper JW, Ryu CM, Zhang SA. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 2004;94:1259-1266
13. Maurhofer M, Hase C, Meuwly P. et al. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology. 1994;84:139-146
14. Loper JE, Hassan KA, Mavrodi DV. et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784
15. Van Loon LC. Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol. 2007;119:243-254
16. Wei ZM, Laby RJ, Zumoff CH. et al. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85-88
17. Baillieul F, Genetet I, Kopp M. et al. A new elicitor of the hypersensitive response in tobacco: A fungal glycoprotein elicits cell death, expression of defense gene, production of salicylic acid, and induction of systemic acquired resistance. Plant J. 1995;8:551-560
18. Ackerveken GFJMVD, Kan JALV, Wit PJGMD. et al. Molecular analysis of the avirulence gene avr9 of the fungal tomato pathogen Cladosporium fulvum fully supports the gene-for-gene-hypothesis. Plant J. 1992;2:359-366
19. Ellis JG, Rafiqi M, Gan P. et al. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr Opin Plant Biol. 2009;12:399-405
20. Arlat M, Gijsegem FV, Huet JC. et al. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 1994;13:543-553
21. De Wit PJGM, Mehrabi R, van den Burg HA. et al. Fungal effector proteins: past, present and future. Mol Plant Pathol. 2009;10:735-747
22. Vivekananthan R, Ravi M, Ramanathan A. et al. Lytic enzymes induced by Pseudomonas fluorescens and other biocontrol organisms mediate defence against the anthracnose pathogen in mango. World J Microb Biot. 2004;20:235-244
23. Acharya K, Chakraborty N, Dutta A K. et al. Signaling role of nitric oxide in the induction of plant defense by exogenous application of abiotic inducers. Archives of Phytopathology and Plant Protection. 2011;44:1501-1511
24. Amelot N, Carrouche A, Danoun S. et al. Cryptogein, a fungal elicitor, remodels the phenylpropanoid metabolism of tobacco cell suspension cultures in a calcium-dependent manner. Plant Cell Environ. 2011;34:149-161
25. Dangl JL, Dietrich RA, Richberg MH. Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell. 1996;8:1793-1807
26. Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205-227
27. Van Loon LC, Bakker PAHM, van der Heijdt WHW. et al. Early Responses of Tobacco Suspension Cells to Rhizobacterial Elicitors of Induced Systemic Resistance. Mol Plant Microbe Interact. 2008;21:1609-1621
28. Kulik A, Noirot E, Grandperret V. et al. Interplays between nitric oxide and reactive oxygen species in cryptogein signalling. Plant Cell Environ. 2015;38:331-348
29. Hammond-Kosack KE, Parker JE. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotech. 2003;14:177-193
30. Atkinson MM, Keppler LD, Orlandi EW. et al. Involvement of plasma membrane calcium influx in bacterial induction of the K +/H + and hypersensitive responses in tobacco. Plant Physiol. 1990;92:215-221
31. Bradley DJ, Kjellbom P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21-30
32. Huang CJ, Tsay JF, Chang SY. et al. Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag Sci. 2012;68:1306-1310
33. Ryu CM, Farag MA, Hu CH. et al. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017-1026
34. Ongena M, Jourdan E, Adam A. et al. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol. 2007;9:1084-1090
35. Garcia-Brugger A, Lamotte O, Vandelle E. et al. Early signaling events induced by elicitors of plant Defenses. Mol Plant Microbe Interact. 2006;19:711-724
36. Pontier D, Godiard L, Marco Y. et al. hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 1994;5:507-521
37. Frias M, Gonzalez C, Brito N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011;192:483-495
38. Chen M, Zeng H, Qiu D. et al. Purification and Characterization of a Novel Hypersensitive Response-Inducing Elicitor from Magnaporthe oryzae that Triggers Defense Response in Rice. Plos One. 2012;7:e37654
39. Hano C, Addi M, Fliniaux O. et al. Molecular characterization of cell death induced by a compatible interaction between Fusarium oxysporum f. sp linii and flax (Linum usitatissimum) cells. Plant Physiol Bioch. 2008;46(5):590-600
40. Schmittgen Thomas D, Kenneth J. Livak. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008:1101-1108
41. Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Mol Bio. 1997;48:251-275
42. Boka K, rban N. New Aspect of H2O2 Signaling. Plant Signal Behav. 2007;2:498-500
43. Noirot E, Der C, Lherminier J. et al. Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J Exp Bot. 2014;65:5011-5022
44. Wang H, Yang X, Guo L. et al. PeBL1, a Novel Protein Elicitor from Brevibacillus laterosporus Strain A60, Activates Defense Responses and Systemic Resistance in Nicotiana benthamiana. Appl Environ Microb. 2015;81:2706-2716
45. Dong XN. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547-552
46. Tjalsma H, Antelmann H, Jongbloed JDH. et al. Proteomics of protein secretion by Bacillus subtilis: Separating the "secrets" of the secretome. Microbiol Mol Biol R. 2004;68:207-233
47. Chen XH, Koumoutsi A, Scholz R. et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007-1014
48. Mur LAJ, Kenton P, Lloyd AJ. et al. The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot. 2008;59:501-520
49. Pontier D, Balague C, Bezombes-Marion I. et al. Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 2001;26:495-507
50. Faoro F, Maffi D, Cantu D. et al. Chemical-induced resistance against powdery mildew in barley: the effects of chitosan and benzothiadiazole. Biocontrol. 2008;53:387-401
51. Torres MA, Jones JDG, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141(2):373-378
52. Angel Torres M. ROS in biotic interactions. Physiol Plantarum. 2010;138:414-429
53. Srivastava N, Gonugunta VK, Puli MR. et al. Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativum. Planta. 2009;229:757-765
54. Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935-944
55. Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiol. 2008;146:839-844
56. Kinkema M, Fan WH, Dong XN. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339-2350
57. Kim HS, Delaney TP. Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J. 2002;32:151-163
58. Zhou JM, Trifa Y, Silva H. et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact. 2000;13:191-202
59. Chaerle L, Lenk S, Hagenbeek D. et al. Multicolor fluorescence imaging for early detection of the hypersensitive reaction to tobacco mosaic virus. J Plant Physiol. 2007;164:253-262
60. Nicholson R L, Hammerschmidt R. Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 1992;30:369-389
Author contact
![]() Corresponding author: Dewen Qiu. E-mail address: qiudewencn; Telephone/Fax: +86 10 82105924
Corresponding author: Dewen Qiu. E-mail address: qiudewencn; Telephone/Fax: +86 10 82105924

 Global reach, higher impact
Global reach, higher impact