10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(9):1074-1082. doi:10.7150/ijbs.15589 This issue Cite
Research Paper
Analysis of PBase Binding Profile Indicates an Insertion Target Selection Mechanism Dependent on TTAA, But Not Transcriptional Activity
1. State Key Laboratory of Genetic Engineering and National Center for International Research of Development and Disease, Fudan-Yale Center for Biomedical Research, Innovation Center for International Cooperation of Genetics and Development, Institute of Developmental Biology and Molecular Medicine, School of Life Sciences, Fudan University, Shanghai 200433
2. Howard Hughes Medical Institute, Department of Genetics, Yale University School of Medicine, New Haven, CT 06536
3. Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan 650500, China
Abstract
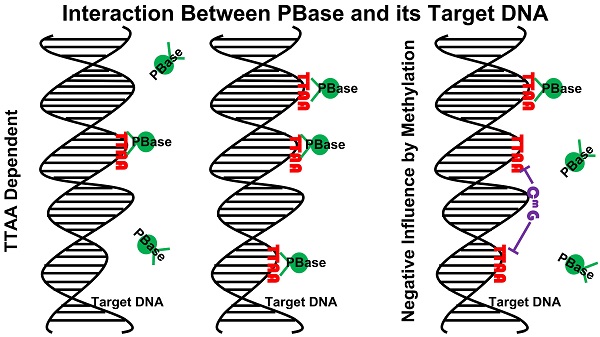
Transposons and retroviruses are important pathogenic agents and tools for mutagenesis and transgenesis. Insertion target selection is a key feature for a given transposon or retrovirus. The piggyBac (PB) transposon is highly active in mice and human cells, which has a much better genome-wide distribution compared to the retrovirus and P-element. However, the underlying reason is not clear. Utilizing a tagged functional PB transposase (PBase), we were able to conduct genome-wide profiling for PBase binding sites in the mouse genome. We have shown that PBase binding mainly depends on the distribution of the tetranucleotide TTAA, which is not affected by the presence of PB DNA. Furthermore, PBase binding is negatively influenced by the methylation of CG sites in the genome. Analysis of a large collection of PB insertions in mice has revealed an insertion profile similar to the PBase binding profile. Interestingly, this profile is not correlated with transcriptional active genes in the genome or transcriptionally active regions within a transcriptional unit. This differs from what has been previously shown for P-element and retroviruses insertions. Our study provides an explanation for PB's genome-wide insertion distribution and also suggests that PB target selection relies on a new mechanism independent of active transcription and open chromatin structure.
Keywords: Transposons, insertion

 Global reach, higher impact
Global reach, higher impact