10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(9):1074-1082. doi:10.7150/ijbs.15589 This issue Cite
Research Paper
Analysis of PBase Binding Profile Indicates an Insertion Target Selection Mechanism Dependent on TTAA, But Not Transcriptional Activity
1. State Key Laboratory of Genetic Engineering and National Center for International Research of Development and Disease, Fudan-Yale Center for Biomedical Research, Innovation Center for International Cooperation of Genetics and Development, Institute of Developmental Biology and Molecular Medicine, School of Life Sciences, Fudan University, Shanghai 200433
2. Howard Hughes Medical Institute, Department of Genetics, Yale University School of Medicine, New Haven, CT 06536
3. Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan 650500, China
Received 2016-3-20; Accepted 2016-5-15; Published 2016-7-18
Abstract
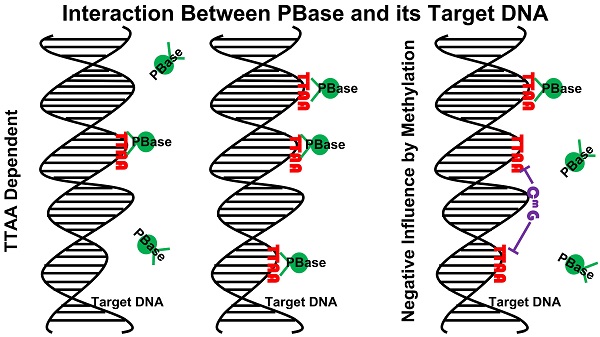
Transposons and retroviruses are important pathogenic agents and tools for mutagenesis and transgenesis. Insertion target selection is a key feature for a given transposon or retrovirus. The piggyBac (PB) transposon is highly active in mice and human cells, which has a much better genome-wide distribution compared to the retrovirus and P-element. However, the underlying reason is not clear. Utilizing a tagged functional PB transposase (PBase), we were able to conduct genome-wide profiling for PBase binding sites in the mouse genome. We have shown that PBase binding mainly depends on the distribution of the tetranucleotide TTAA, which is not affected by the presence of PB DNA. Furthermore, PBase binding is negatively influenced by the methylation of CG sites in the genome. Analysis of a large collection of PB insertions in mice has revealed an insertion profile similar to the PBase binding profile. Interestingly, this profile is not correlated with transcriptional active genes in the genome or transcriptionally active regions within a transcriptional unit. This differs from what has been previously shown for P-element and retroviruses insertions. Our study provides an explanation for PB's genome-wide insertion distribution and also suggests that PB target selection relies on a new mechanism independent of active transcription and open chromatin structure.
Keywords: Transposons, insertion
Introduction
Insertion profiles are key feature of transposons and retroviruses which can aid in increasing our understanding of pathogenesis and in developing tools for mutagenesis and transgenesis. It has been reported that both transposons and retroviruses preferably insert into transcriptionally active genes, which may be due to the higher accessibility of open chromatin and/or the interaction between transposase/integrase and cellular proteins bound to transcriptionally active regions (1-6). The distribution of P-element insertions in the Drosophila melanogaster genome exhibits high preference for transcriptionally active regions, and especially for the regions near the transcriptional start site (7, 8). A similar preference has been also reported for retroviruses including human immunodeficiency virus (HIV), avian sarcoma-leukosis virus (ASLV), and murine leukaemia virus (MLV) (2, 3, 9-13). It has been shown that transcriptional co-activators bound to HIV integration complexes serve as a tether for insertion targeting (14-17).
The piggyBac (PB) transposon has been shown to be highly active in mice and human cells (18), making it an ideal tool for a variety of genetic manipulations in mammals and human cells (18-28). In fact PB has many advantageous features including having the highest transposition efficiency compared to other DNA transposons (29, 30), being active in broad cell types and species from insects to mammalians (18-20, 31-36), capable of carrying a large cargo (23), and precise excision without leaving damage footprints (18, 37, 38). PB appears to have a preference for transcriptional units (18, 21, 22), suggesting that it could also have a mechanism similar to P-element and retroviruses that target transcriptionally active genes. On the other hand, PB has a significantly broader genome-wide distribution than other transposons and retroviruses (30, 39, 40), arguing that it may employ a different mechanism in target selection.
To better understand the genomic profile of PB insertion site selection, we have tagged the PB transposase (PBase) and examined PBase binding preference in the mouse genome using the Chromatin Immunoprecipitation (ChIP)-Chip method (41, 42) which is a high-throughput method used to identify the interaction between genomic DNA and the protein of interest in the living cells. We have analyzed the PBase binding distribution in the genome in relation to the distribution of PB's target sequence TTAA, the distribution of transcriptional units, gene expression levels, and the distribution of CG methylation. Our analysis has revealed that unlike P-element and retroviruses, PBase binding does not prefer transcriptional active units, but rather mainly depends on the distribution of TTAA sites with a negative influence by CG methylation. This unique mechanism provides an explanation for the behavior of PB's genome-wide insertion profile and broad activities in different species and cell types.
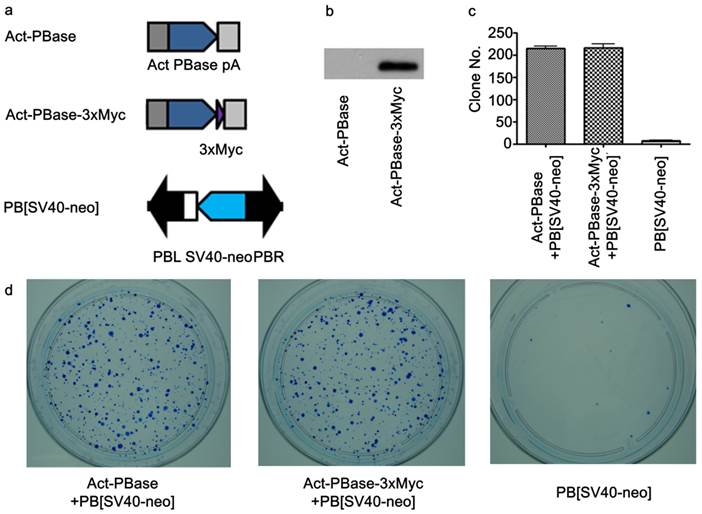
Utilizing Myc-tagged PBase for transposition in cultured mouse ES cells. (a) Both PBase and PBase-3×Myc were driven by an actin promoter. Tri-Myc tag (Blue triangle) was added to the c-terminal of PBase (PBase-3×Myc). PB [SV40-neo] carrying the neo drug selection marker driven by a SV40 promoter served as the donor plasmid (21). (b) Transient expression of PBase-3×Myc in ES cells 48h after transfection. (c) Statistical results of PB transposition efficiency test (21). PBase-3×Myc drove transposition with the same efficiency as PBase (p=0.6). Each number is the average obtained from three experiments. (d) An example of ES cell transposition efficiency test experiments. Blue dots were surviving ES clones stained by methylene blue after G418 selection. The number of survival cell clones after G418 selection indicated the transposition efficiency.
Results
Mapping of PBase binding sites in mouse ES cells
To investigate interactions between PBase and the mouse genomic DNA, we applied the ChIP-Chip method using the Affymetrix® Chromatin Immunoprecipitation Assay Protocol (P/N 702238). 3×Myc-tag were added to the C-terminal of PBase (Figure 1a). This Myc tagged PBase was shown to be (Figure 1b) and able to catalyze PB transposition in W4/129S6 mouse ES cell (Taconic) genome with the same efficiency as wild-type PBase (Figure 1c, 1d). 15μg Myc antibody (SC-789X, Santa Cruz) was used to immunoprecipitate the PBase-DNA complex. The Affymetrix® GeneChip® Mouse Tiling 2.0R Array Set (P/N 900852) was then used to identify PBase binding sites. To check whether the PB transposon had an effect on PBase binding, we mapped the PBase binding sites in ES cells with and without the presence of the PB transposon designated as PB+ or PB-, respectively. For experiments with each group, 2 replicates were performed. Using the peak finding algorithm (43, 44), we defined PBase binding sites across the whole mouse genome. These sites displayed similar distribution patterns in all the analyses carried out for this study. We did not detect any effect of the PB transposon on the distribution of the PBase binding sites in our study. We then aggregate all the sites and identified a total of 2,396,017 PBase binding sites across the mouse genome (Table S1).
TTAA is highly enriched at the center of PBase binding sites
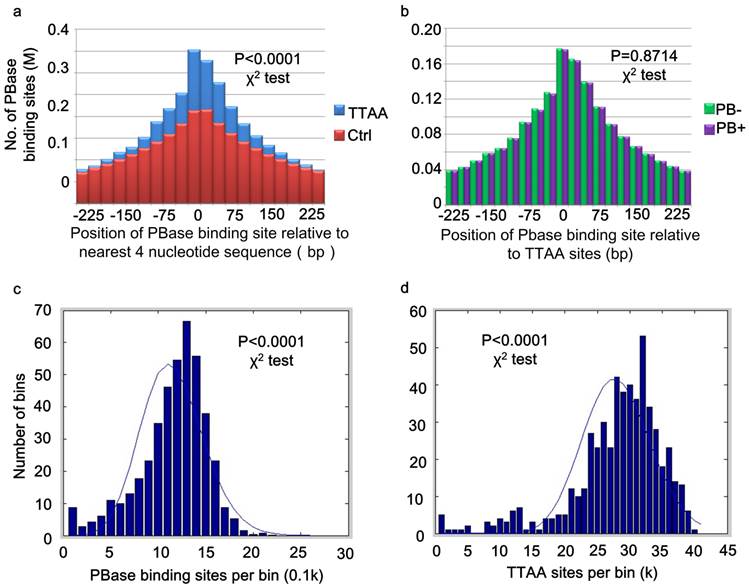
PB inserts almost exclusively into TTAA sites (37, 45). But little is known about the mechanism of target determination. We matched our 2,396,017 PBase binding sites to the TTAA sites across the genome. 87.85% (2,104,819) of these binding sites were within 250 bp of the TTAA sites and 25.48% (610,405) of the binding sites were within 25 bp. The TTAA motif was significantly enriched at the center of binding sites compared to the other combinations of 4 nucleotides (χ2 analysis, p < 0.0001) (Figure 2a).
For those peaks located within 25bp of TTAA sites, 51.21% (306471) were from the PB- group, while 49.79% (303934) were from PB+ group. There was no significant difference between the distribution of binding sites from PB+ and PB- group relative to TTAA sites (χ2 analysis, p =0.8714) (Figure 2b). These data indicated that even without the presence of PB transposon, PBase already has the ability to bind TTAA sites.
TTAA sites were enriched at the center of PBase binding sites. (a) Distribution of PBase binding sites relative to the TTAA sites. Other combination of four nucleotides sequences served as control. (b) TTAA was enriched at the center of PBase binding sites even without the presence of PB transposon. (c) The non-random distribution of both PBase binding sites and (d) TTAA sites in the mouse genome. Mouse genome was divided into 5Mb bins. The number of PBase binding sites or TTAA sites in each bin was calculated. Both distributions were significantly different from Possion distribution (χ2 test, P < 0.0001).
PBase binding site distribution follows that of TTAA sites in the genome, but not gene density or transcriptional activity levels
Global analysis of PBase binding sites showed that the sites spread universally across the whole mouse genome, but are not strictly randomly (Poisson) distributed (χ2 analysis, p < 0.0001) (Figure 2c). This might be partially due to the non-random distribution of TTAA sites in the mouse genome (χ2 analysis, p<0.0001) (Figure 2d). The distribution of PB insertion sites during large-scale mutagenesis in the D. melanogaster genome is also not random (8). We found that TTAA sites are also not randomly distributed in the D. melanogaster genome either (χ2 analysis, p < 0.0001).
Further analysis of the distribution of PBase binding sites in the mouse genome showed that the PBase binding sites had no preference for any particular chromosome. The number of binding sites on each chromosome was highly correlated with the chromosome length (CC = 0.9800) and TTAA density (CC=0.9682) (Figure 3a), but not correlated with the gene density (CC=0.1613). These data suggest that unlike P-element and retroviruses, PBase binding followed TTAA, but not transcription activity.
Global distribution of PBase binding sites followed that of TTAA sites. (a) The distribution of PBase binding sites in the whole mouse genome at the chromosome level. The density of PBase binding sites on each chromosome was highly correlated with the length of this chromosome and the density of TTAA sites. (b) The distribution of PBase binding sites or (c) PB insertion sites in different transcription element was shown. High correlation was detected between the density of PBase binding sites on a gene and (d) the density of TTAA sites or (e) the length of this gene. (f) Expression level of a gene had no correlation with PBase binding. BT: binding sites to TTAA; BL: binding sites to length; IT: insertion sites to TTAA; IL: insertion sites to length.
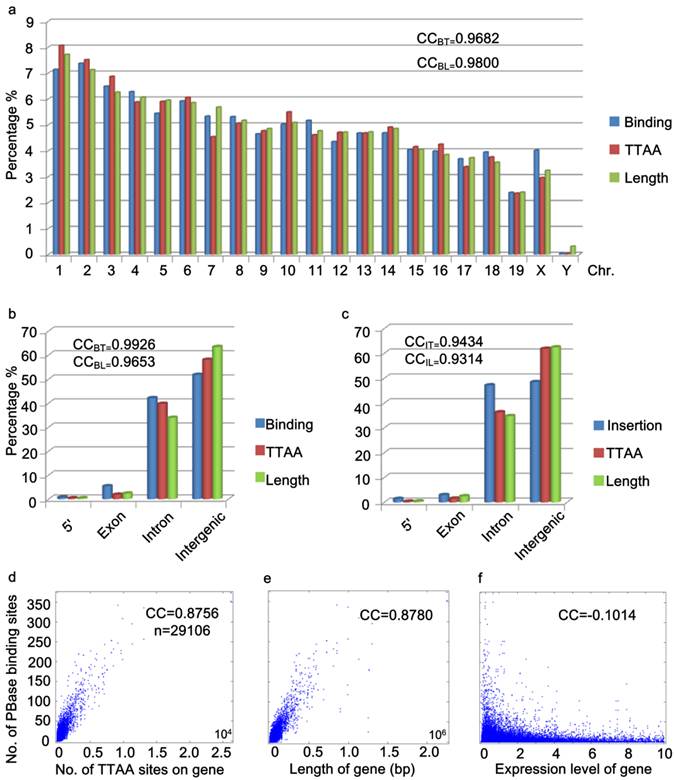
To further explore the potential relationship between PBase binding and transcriptional activity, we performed two more analyses. Previously, P-element and retrovirus insertions were shown to exhibit a strong preference for the 5' region upstream of the transcription start sites (2, 3, 7-13, 40, 46). We did not detect a strong 5' upstream preference for PBase binding. Our data showed that only 0.72% of PBase binding sites were located in the 5' region within 1000bp upstream of transcription start sites (Figure 3b). Unlike the dramatically distorted distributions of P-element insertions (73%) in the D. melanogaster genome (40) and retroviruses (8%-20%) in the human genome (46), the distribution of PBase binding sites in the 5' region within 1000bp upstream of the transcription start site, exonic and intronic regions (5': 072%, exon: 5.36%, intron: 42.11%) is largely correlated with the length (5': 0.37%, exon: 2.36%, intron: 33.89%, CC=0.9653) or the TTAA density (5': 0.37%, exon: 1.86%, intron: 39.73%, CC=0.9926) of these regions in mouse genome (Figure 3b). Similar distribution patterns were also observed by analyzing our 5248 PB insertion sites in mouse mutant strains generated in germline transposition experiments (5': 1.39%; exon: 2.93%, intron: 47.18%, CC=0.9434) (Figure 3c).
The strong 5' preference of P-element and retrovirus insertion has been attributed to the accessibility of the open chromatin regions that correlate with transcriptional activity (4, 9). Our data for PB suggest that PB does not rely on transcriptional activity. We thus checked whether transcription levels affect PBase binding in the mouse genome. We obtained the microarray expression data of 29106 transcripts from the mouse ES cells (47). No correlation between the expression level of the transcripts and the density of PBase binding sites were detected (CC = - 0.1014) (Figure 3f). On the other hand, a strong correlation between the number of TTAA sites in a gene (or the length of a gene) and the density of PBase binding sites on a gene was detected (CC = 0.8756 / 0.8780) (Figure 3d, 3e). Together these results indicate that PBase binding would largely depends on the TTAA sites rather than a region's transcriptional activity status.
Distribution of PBase Binding Is influenced by Methylation of CpG
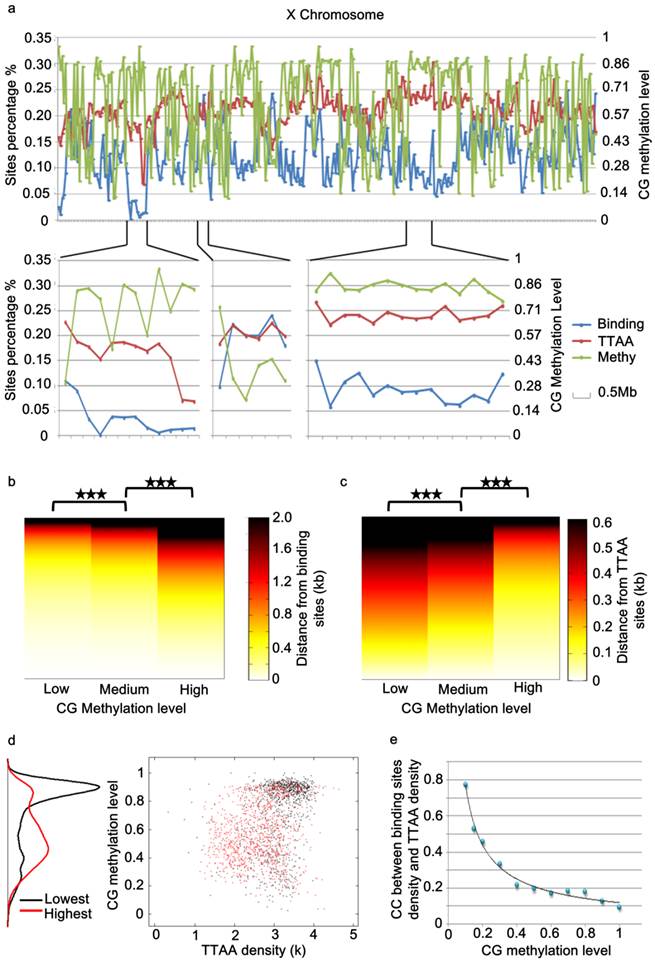
While the global distribution of PBase binding sites generally followed that of TTAA sites in the genome, we noticed that there are regional distortions of this distribution (Figure 4a), suggesting other local factors might influence PBase binding besides TTAA. Further analysis showed that those regions, in which the density of PBase binding sites was much lower and not in proportion to TTAA density, were most probably highly methylated CG regions (Figure 4a, 4d). These trends suggested that CG methylation has a negative influence on PBase binding. To confirm this, we divided all the CG sites into 3 groups according to methylation levels. Those highly methylated CG sites were significantly farther away from PBase binding sites compared to low methylated sites (t test, p<0.001)(Figure 4b), which indicates that highly methylated CG sites prevent PBase from binding nearby. This phenomenon was not an artifact of TTAA distribution because TTAA sites have an opposite distribution trend in comparison to CG sites (Figure 4c). Consistent with this, when we only analyzed the low methylated CG regions (methylation level < 0.1), the density of PBase binding sites was correlated with the density of TTAA sites (CC=0.7741) (Figure 4e). These observations together indicate that the CG methylation of target DNA have an inhibitory effect on PBase binding.
Discussion
It has been proposed that DNA transposons and retrovirus integration in the genome depends on active transcription. The piggyBac transposon has been shown previously to target TTAA motif for integration. However, it is not clear whether the PB transposon has a preference for TTAA sites in actively transcribed regions in the genome. To define the target selection profile for piggyBac, we introduced a Myc-tagged and functional PBase into the mouse ES cells and were able to generate a genome-wide map of PBase binding sites in the mouse genome. We found that PBase binding sites spread evenly in the mouse genome depending on the distribution of TTAA sites, but did not correlate with gene density or gene expression levels. The insertion sites from a large germline mutagenesis screen are consistent with the results of our PBase binding site analysis. While retroviruses and P-element have a strong bias for transcriptionally active regions, the PBase binding profile showed that PBase binding selection does not prefer transcriptionally active regions, but rather largely depends on the distribution patterns of TTAA sites. Our data therefore suggest a transposition mechanism different from retroviruses and P-element. It has been shown that integrases from retroviruses interact with cellular proteins or co-factors that are bound to transcriptionally active regions. This could permit these integrases have a higher accessibility to open chromatins or transcriptionally active regions. The lack of preference for transcriptionally active regions by PBase suggests that it might not interact with cellular proteins that bind to transcriptionally active regions. Furthermore, TTAA is a short sequence and is widely distributed throughout the genomes of different organisms. This could explain the broader genome-wide distribution patterns of PB and its capacity to transpose in a wide range of hosts. PB's unique target selection profile makes it an ideal tool for genome-wide mutagenesis.
PBase binding was inhibited by the methylation of genomic DNA. (a) A chromosome view of the PBase binding sites density, TTAA sites density and CG methylation level of chromosome x. PBase binding was affected in the highly methylated CG regions. (b) Relative distribution of PBase binding sites to CG sites. CG sites were divided into 3 groups based on the methylation level. PBase tended to bind near the low methylated CG sites. This is not caused by the distribution of the TTAA sites, which was an opposite trend (c). (d) Regions with the lowest PBase binding in the whole genome, were more likely high methylated regions (black dots). (e) The correlation coefficient value between PBase binding sites density and TTAA sites density was increased when those highly methylated regions were removed.
We also detected a negative influence of genomic methylation on PB's target selection, which is similar to the previous finding that the methylation of PB transposon DNA itself inhibits transposition (19). Therefore, methylation could be a mechanism that silences PB. While this feature could affect PB mutagenesis for somatic cells or in tissue culture cells, its germline mutagenesis capacity should not be affected as the genome of the germ line cells is largely unmethylated (48).
In summary, PB's distinctive and transcription-independent target selection profile suggests a transposition mechanism different from retroviruses and P-element. Future studies exploring the interaction between the transposases and host factors could provide molecular mechanisms that contribute to this difference.
Materials and Methods
Plasmid Construction
Construction of Act-PBase-3×Myc was as follows: the coding sequence of 3×Myc was PCR amplified from pIND-3×Myc with primer1 (5'-TCAACGAAAGTACCGGTAAACC-3') and primer2 (5'-ATAGTATAGCGGCCGCCTTGTACTCGGAAACAA-3') , and cloned into the Not I and Age I sites of Act-PBase (18) to generate Act-PBase-3×Myc.
Mouse ES Cell Transfection
W4/129S6 mouse ES cells (Taconic) were used for the ChIP-chip assay to detect PBase binding sites in the mouse genome in this study. The conditions for culture and electroporation of ES cells were described in the manufacturer-recommended protocols (Taconic). 30μg circular Act-PBase-3×Myc in PB- group or 30μg Act-PBase-3×Myc plus 5μg PB[SV40-neo](18) in PB+ group were used for electroporation of 107 cells. After electroporation, cells were seeded onto 10 cm dish containing mitomycin C-treated mouse embryonic fibroblast feeder cells, ES cells were then harvested for ChIP after a 48h incubation period.
Transposition efficiency test
20μg circular PB[SV40-neo] and 10μg Act-PBase-3×Myc (or Act-PBase) were used for electroporation. Geneticin (G418) was added into each dish at the final concentration of 500μg/ml for selection neo resistant clones after 48h incubation. The medium was changed every day with geneticin. After 7 days selection, cells were fixed with PBS containing 4% paraformaldehyde for 10 minutes and then stained with 0.2% methylene blue for an hour. Cell clones were counted after washing with deionized water.
ChIP-chip
ES cells were harvested and crosslinked with 1% formaldehyde 48h after transfection. Sonication was performed with Sonics® VC 130 Vibra cellTM at 35 amplitude, 30 seconds pulses with 1 minute rest, 10 cycles to shear the DNA to 100-1000 bp fragments. Both pulsing and resting steps were performed in an ice bath. ChIP-chip experiments were performed according to Affymetrix® Chromatin Immunoprecipitation Assay Protocol (P/N 702238). A total of 108 ES cells were used per immunopreicipitation (IP) with the use of 15μg anti-Myc antibody (SC-789X, Santa Cruz). GeneChip® Mouse Tiling 2.0R Array Set (P/N 900852), a whole mouse genome tiling array was used for the DNA analysis.
Identification of PBase Binding Sites
The CHIP-chip data was first normalized using the quantile normalization. Then the peak finding algorithm (43) was applied to identify PBase binding sites. There are two main steps in this algorithm: First, identification of the binding region. Candidate binding regions should satisfy the following thresholds: (i) should contain at least 4 probes with significantly higher signal intensity than the background; and (ii) The distance between each neighboring probe with significantly higher signal in the region should be no more than 500bp. Second, identification of candidate binding sites from the binding region. For each binding region identified in the previous step, a double standard linear regression was performed which fits neighboring signals to asymmetric triangles centered on candidate binding sites. In total, we defined 2396017 PBase binding sites in the whole mouse genome (Table S1). This program was implemented using the Java programming language.
Supplementary Material
Figure S1 and Table S1.
Acknowledgements
We thank Drs. Xiaohui Wu, Wufan Tao, Rener Xu, Min Han and Yuan Zhuang for discussion and comments. We also acknowledge Mr. Xu Yang, Dr. Lei Yao, Mr. Kunlin Liu and Dr. Xinran Dong for the help with perl scripts. Ms. Boyin Tan is acknowledged for the help in ES cell culture. We want to give special thanks to Prof. Beibei Ying for the encouragement and support. This work is supported by Chinese Key Projects for Basic Research (973) (Grant No.2006CB806700), Hi-tech Research and Development Project (863) (Grant No. 2007AA022012).
Author Contributions
Dong Yang conducted the ChIP-Chip experiment and the global distribution analysis of PBase binding sites. Ruiqi Liao defined the PBase binding sites from chip data. Yun Zheng mainly advised on software for PBase binding sites identification. Ling Sun mainly advised on the ChIP-Chip experiments and data analysis. Tian Xu initiated this project and mainly advised on data analysis.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Molecular and cellular biology. 2005;25(6):2085-94
2. Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521-9
3. Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC. et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS biology. 2004;2(8):E234
4. Panet A, Cedar H. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell. 1977;11(4):933-40
5. Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S. et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. Journal of virology. 2005;79(11):6610-9
6. Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34(3):297-302
7. Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(24):10824-30
8. Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM. et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36(3):283-7
9. Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S. et al. Genome-wide analysis of retroviral DNA integration. Nature reviews Microbiology. 2005;3(11):848-58
10. Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300(5626):1749-51
11. Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(17):6103-7
12. De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E. et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105(6):2307-15
13. Laufs S, Nagy KZ, Giordano FA, Hotz-Wagenblatt A, Zeller WJ, Fruehauf S. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Molecular therapy: the journal of the American Society of Gene Therapy. 2004;10(5):874-81
14. Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M. et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. The EMBO journal. 2001;20(12):3272-81
15. Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88(4):483-92
16. Suzuki Y, Craigie R. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. Journal of virology. 2002;76(23):12376-80
17. Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M. et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. Journal of virology. 2004;78(17):9524-37
18. Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473-83
19. Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D. et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(27):9290-5
20. Wilson MH, Coates CJ, George AL Jr. PiggyBac transposon-mediated gene transfer in human cells. Molecular therapy: the journal of the American Society of Gene Therapy. 2007;15(1):139-45
21. Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM. et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32(8):837-44
22. Landrette SF, Cornett JC, Ni TK, Bosenberg MW, Xu T. piggyBac transposon somatic mutagenesis with an activated reporter and tracker (PB-SMART) for genetic screens in mice. PLoS One. 2011;6(10):e26650
23. Li R, Zhuang Y, Han M, Xu T, Wu X. piggyBac as a high-capacity transgenesis and gene-therapy vector in human cells and mice. Dis Model Mech. 2013;6(3):828-33
24. Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S. et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330(6007):1104-7
25. Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766-70
26. Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771-5
27. Gayle S, Pan Y, Landrette S, Xu T. piggyBac insertional mutagenesis screen identifies a role for nuclear RHOA in human ES cell differentiation. Stem Cell Reports. 2015;4(5):926-38
28. Ni TK, Landrette SF, Bjornson RD, Bosenberg MW, Xu T. Low-copy piggyBac transposon mutagenesis in mice identifies genes driving melanoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(38):E3640-9
29. Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S. et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103(41):15008-13
30. Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P. et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther. 2010;18(10):1803-13
31. Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect biochemistry and molecular biology. 2002;32(10):1211-20
32. Sumitani M, Yamamoto DS, Oishi K, Lee JM, Hatakeyama M. Germline transformation of the sawfly, Athalia rosae (Hymenoptera: Symphyta), mediated by a piggyBac-derived vector. Insect biochemistry and molecular biology. 2003;33(4):449-58
33. Lorenzen MD, Berghammer AJ, Brown SJ, Denell RE, Klingler M, Beeman RW. piggyBac-mediated germline transformation in the beetle Tribolium castaneum. Insect molecular biology. 2003;12(5):433-40
34. Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito Anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. The Journal of biological chemistry. 2002;277(11):8759-62
35. Gonzalez-Estevez C, Momose T, Gehring WJ, Salo E. Transgenic planarian lines obtained by electroporation using transposon-derived vectors and an eye-specific GFP marker. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14046-51
36. Park TS, Han JY. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(24):9337-41
37. Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect molecular biology. 1996;5(2):141-51
38. Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98(1):33-41
39. Meir YJ, Weirauch MT, Yang HS, Chung PC, Yu RK, Wu SC. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011;11:28
40. Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E. et al. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188(3):731-43
41. Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I. et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306-9
42. Zheng M, Barrera LO, Ren B, Wu YN. ChIP-chip: data, model, and analysis. Biometrics. 2007;63(3):787-96
43. Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA. et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436(7052):876-80
44. Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ. et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18(1):46-59
45. Fraser MJ, Cary L, Boonvisudhi K, Wang HG. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995;211(2):397-407
46. Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C. et al. Genome-wide analyses of avian sarcoma virus integration sites. Journal of virology. 2004;78(21):11656-63
47. Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, Campbell PA. et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85
48. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766-70
Author contact
![]() Corresponding authors: Ling Sun (lingsunedu.cn) and Tian Xu (tian.xuedu)
Corresponding authors: Ling Sun (lingsunedu.cn) and Tian Xu (tian.xuedu)

 Global reach, higher impact
Global reach, higher impact