Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(8):1092-1099. doi:10.7150/ijbs.21206 This issue Cite
Review
Structural Maintenance of Chromosomes protein 1: Role in Genome Stability and Tumorigenesis
Key Laboratory of Medical Cell Biology, Ministry of Education; Institute of Translational Medicine, China Medical University; Liaoning Province Collaborative Innovation Center of Aging Related Disease Diagnosis and Treatment and Prevention, Shenyang, Liaoning Province, China
Received 2017-5-27; Accepted 2017-7-5; Published 2017-9-3
Abstract
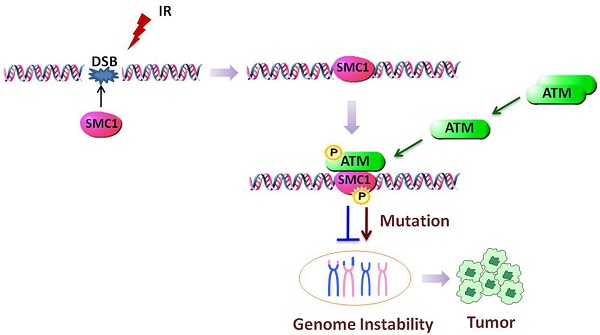
SMC1 (Structural Maintenance of Chromosomes protein 1), well known as one of the SMC superfamily members, has been explored to function in many activities including chromosome dynamics, cell cycle checkpoint, DNA damage repair and genome stability. Upon being properly assembled as part of cohesin, SMC1 can be phosphorylated by ATM and mediate downstream DNA damage repair after ionizing irradiation. Abnormal gene expression or mutation of SMC1 can cause defect in the DNA damage repair pathway, which has been strongly associated with tumorigenesis. Here we focus to discuss SMC1's role in genome stability maintenance and tumorigenesis. Deciphering the underlying molecular mechanism can provide insight into novel strategies for cancer treatment.
Keywords: SMC1, phosphorylation, cell cycle regulation, genome stability, DNA damage repair, tumorigenesis
Introduction
As a member of a subfamily chromosome-associated proteins known as SMCs (Structural Maintenance of Chromosomes), which are highly conserved from bacteria to humans, SMC1 is a core component of the cohesin complex that is required for sister chromatid cohesion [1-3]. In addition, SMC1 is involved in a variety of cellular functions, including cell survival [4], chromosome dynamics [5-7], cell cycle regulation [8, 9] and DNA damage repair [10-12]. Importantly, SMC1-mediated chromosome structure stability and DNA damage repair are considered as important mechanisms for the maintenance of genome integrity. Although there is increasing evidence that SMC1 is closely associated with various cancer types, current knowledge about its role in tumorigenesis remains limited.
In this review, we summarize the current knowledge on SMC1 in cell cycle regulation, genomic stability and its mutations in various cancers, and discuss the potential role of SMC1 serving as a biomarker in clinical cancer diagnosis.
Overview of SMC1
The SMC proteins were initially identified through genetic studies of chromosome segregation in Saccharomyces cerevisiae [13]. Smc1p, characterized as the founding member of SMC1 family proteins by frequent minichromosome nondisjunction in mutations [14], was later shown to be essential for viability and maintaining cohesion between sister chromatid [13].
The SMC1 protein contains a N-terminus ATP binding domain and a C-terminus ATP hydrolysis domain, which are separated by two long coiled-coils of 200-450 residues and a central globular hinge region [15, 16]. In an antiparallel orientation, these two domains form a functional ATPase [17-19], which could regulate DNA binding and tethering [20, 21]. Furthermore, deacetylation of SMC1 and SMC3 dimers promotes dissociation of the coiled- coil arms [22].
Classically, SMC1 and SMC3 are believed to form a heterodimer in an antiparallel mode as the core of cohesin complex, which is required for sister chromatid cohesion during replication [1, 2, 23, 24], and is also involved in recombination as part of the RC-1 complex with DNA polymerase ε and ligase Ⅲ [3, 25, 26]. In humans, two different isoforms of SMC1, SMC1A and SMC1B have been discovered. SMC1A is the core subunit of the mitotic cohesin complex, which is composed of SMC1A, SMC3, Rad21 [27-30] and SA1 or SA2 [3, 28, 31, 32]. SMC1B is currently thought as a meiotic specific subunit of cohesin complex, called SMC1B, REC8 or STAG3 respectively [33], which plays a key role in sister chromatid pairing and preventing telomere shortening [34-36]. Recent studies showed that SMC1B is also expressed in somatic mammalian cells as a member of a mitotic cohesin complex preserving genome stability in response to irradiation [37].
SMC1 in chromosome dynamics and cell cycle
Under normal conditions, SMC1 as part of cohesin, is recruited to chromatin in G1 phase with sister chromatid cohesion taking place in S phase, and holds sister chromatid together until accurate chromosome segregation in M phase during the cell cycle [38-41]. In addition, SMC1 was also found to be present at centrioles of a centrosome at G0/G1 stage to function as a centrosomal protein [42]. During mitosis, SMC1 was observed at the spindle poles [43, 44] and the active centromere on dicentric chromosomes in concomitant with functional kinetochore [45]. It was reported that the injection of an antibody specific for hSMC1 into human mitotic cells blocked the progression of metaphase and led to disorganization of the metaphase plate [46]. Cytoplasmic mislocation of SMC3 and degradation of SMC1 and RAD21 can also be induced by SMC1 knockdown [47]. These results demonstrate that SMC1 is tightly associated with cell cycle progression and may function as a unique regulator.
SMC1 participates in DNA damage repair and genomic stability maintenance
In addition to the canonical role in sister chromatid cohesion, SMC1 is also a core component of the tetrameric complex cohesin, which coordinates the HR (Homologous Recombination) pathways for DNA DSB (Double Strand Breaks) repair [48, 49]. SMC1 deletion was found to compromise the DSB repair in G2-phase cells [50], but phosphorylation of SMC1 is actually the final performer of its function. Yazdi and Kim et al. firstly reported that SMC1, which is phosphorylated by ATM in serine 957 and 966, acts as a downstream effector of a separate branch of the S-phase checkpoint pathway, namely the ATM/NBS1/SMC1 pathway in response to IR (Ionization Radiation) [51-53]. This pathway could also be triggered by H/R (Hypoxia and Reoxygenation)-induced DNA damage [54]. In this pathway, both NBS1 and BRCA1 were required for the recruitment and maintenance of activated ATM to the site of DSB associated with efficient phosphorylation of SMC1 by ATM [52], which could be disrupted by the UBD (Ubiquitin Binding Domains) of RAD18 [55]. SMC1 could also be phosphorylated after exposure to UV irradiation or hydroxyurea by ATR as well [56-59]. Furthermore, the absence of SMC1A leads to chromosomal aneuploidy and aberrations at fragile sites in human fibroblast cells [57]. Cells expressing phosphorylation mutant of SMC1 show a defective S-phase checkpoint, decreased survival and increased chromosomal aberrations after IR [60]. However, phosphorylation of SMC1 dose not alter the IR-induced G2/M checkpoint, while having an effect on G2 phase accumulation [51].
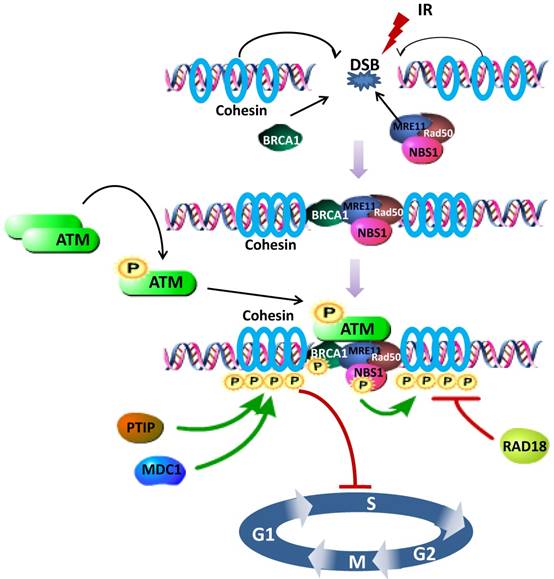
Increasing evidence demonstrates that the fully assembled cohesin is required for phosphorylation of SMC1, in other words, SMC1 is phosphorylated as part of cohesin [11, 61]. Specifically, SMC1 phosphorylation requires the phosphorylation of NBS1 by ATM, establishing NBS1, whose N- and C- terminus are indispensable for the interaction with SMC1, as an adaptor in the ATM/NBS1/SMC1 pathway [51]. NBS1-T278 phosphorylation may be a major site assisting SMC1 phosphorylation upon high dose of DNA damage [62]. Meanwhile, absence of NBS1 causes delay of SMC1 phosphorylation [63]. As either regulators or cofactors, several other proteins are also uncovered to be involved in regulating the SMC1 pathways for cell cycle checkpoint and DNA damage repair. For example, knocking down MDC1 (Mediator of DNA Damage Checkpoint protein 1) decreases the phosphorylation of SMC1 [64] and prevents activation of the intra-S phase and G2/M phase cell-cycle checkpoints after exposure to IR. PTIP (Pax2 transactivation domain-interacting protein) upstreams 53BP1-ATM-SMC1 pathway, where SMC1 is phosphorylated at the DNA damage sites, for DNA DSBs repair and activation of the DNA damage-induced intra-S phase checkpoint [65] (Figure 1).
In addition, SMC1 participates in the maintenance of genomic stability and is required for bipolar mitosis in human cells [66]. ATM anchored at spindle pole or a related kinase presumably phosphorylates Ser-957 and Ser-966 of SMC1, which in turn stimulates its binding to the microtubule-associated Rae1 [44]. Overexpression of wild-type SMC1, but not the mutant SMC1-S957A, SMC1-S966A or SMC1-DBA (Double S957A and S966A) that fails to interact with Rae1, resulted in more multipolar and less bipolar spindle phenotypes [44, 67].
Mouse models with SMC1A targeted showed a series of phenotypic characteristics in development. MGI (Mouse Genome Informatics) database provided the valuable information in targeted mutation mouse of SMC1AS957AS966A (SMC1Atm1Mbk, tm1: targeted mutation 1; Mbk: Michael B Kastan). The phenotype of SMC1Atm1Mbk mouse were premature death, abnormal hematopoietic and immune system.
Nevertheless, it is generally believed that SMC1B is a meiotic specific member of cohesin. The current study showed that SMC1B was also expressed in somatic mammalian cells. The cell cycle progression of SMC1B-slienced cells was alike to the control cells. SMC1B was shown to safeguard genome stability by allowing DSB repair following IR whereas its depletion had no effect on chromosome cohesion [37]. Interestingly, the phenotypes of SMC1B-deficient mice were different. Initial observations of mice deficient in SMC1B were conducted by Revenkova et al. They found that SMC1B-/- mice were viable and showed normal mating behavior. However, the weight of SMC1B-/- mice was only half of heterozygous or wild-type littermates. Both males and females were infertile in SMC1B-/- mice. Males exhibited meiotic arrest in pachytene. Female meiosis progressed through meiosis I until metaphase II, but premature loss of sister chromatid cohesion resulting in massive aneuploidy [7]. Furthermore, SMC1B-/- female mice showed shifting of chiasmata placement and missing of cohesion with age [68]. In summary, SMC1-mediated DNA damage repair and genomic stability maintenance are prevalent under conditions of IR or UV radiation, and this process can be modulated by its phosphorylation. Thus, it is quite clear that alteration of expression and phosphorylation mutations could cause dysfunction of cell cycle checkpoint and failure of DNA damage repair which lead to genomic instability and tumorigenesis.
Schematic representation of the SMC1 participating in DNA damage repair signaling pathway. After DSB induced, BRCA1, cohesin (SMC1, SMC3 and RAD21) and MRN complex (MRE11, Rad50 and NBS1) are recruited to the damage site. Intermolecular autophosphorylation of ATM dimmers leads to release of active ATM monomers. Active ATM monomers can phosphorylate substrates such as BRCA1, NBS1 and SMC1. Both BRCA1 and NBS1 are required for the phosphorylation of SMC1, fully assembled as part of cohesin, by activated ATM, thus inducing S phase arrest. PTIP, MDC1 and RAD18 are involved in regulating SMC1 phosphorylation. Green Arrows represent activating events and red perpendicular ends represent inhibitory events.
SMC1 and its mutations in tumorigenesis
The role of SMC1 in cancer
SMC1, whose abnormality has been enumerated as enabling characteristics of cancer, properly appears to contribute to chromosome stability, DNA damage checkpoint and repair [69]. Thus it is plausible to suggest that the expression of SMC1 and its mutations are able to influence cells from being normal to malignant. SMC1 has been previously found to be involved in the pathogenesis of several types of tumors associated with up-regulation of its expression. SMC1A is over expressed in colorectal cancer tissues and its expression level is correlated with poor prognosis for late stage patients. Increased expression of SMC1A is also profoundly associated with distant metastasis and advanced TNM (Tumor Lymph Nodes Metastasis) stage of disease. In contrast, SMC1A knockdown resulted in a significant inhibition of cell proliferation, colony formation, cell cycle progression and up-regulation of apoptosis in colorectal cancer cells, as well as prevention of colon tumorigenesis in nude mouse models [70]. Accordingly, SMC1A deficiency results in inhibition of cell proliferation, increased apoptosis and enhanced chemosensitivity to oxaliplatin [71]. Being consistent with its role in tumorigenesis, SMC1A was found to be highly expressed in TNBC (Triple-Negative Breast Cancer) cell lines, localizing in the nucleus, cytoplasm and plasma membrane along with SMC3 [72]. Overexpression of SMC1A also increases EMT (Epithelial-Mesenchymal Transition) in TNBC through the induction of Brachyury, a protein encoded by the T gene in humans, which is a transcription factor within the T-box complex of genes [73]. Moreover, SMC1A is highly expressed in human glioma tissues versus surrounding normal tissues and U25L cells, while knocking down SMC1A expression leads to G2/M arrest, decreased cell growth and cyclin B1 expression [74, 75]. SMC1B expression is closely related to increased invasiveness and aggressiveness of several cancers [76-78]. High SMC1B expression has been shown to be associated with pancreatic cancer [76] and the radioresistant H1299 cells than in the radiosensitive H460 cells [78].
Conversely, in other types of tumors, such as AML (Acute Myelocytic Leukemia), patients, with a moderate to high SMC1A protein expression, show a significantly improved cancer-free survival as well as overall survival, which suggests that low expression of SMC1A protein could have a poor prognosis [79]. However, in patients with early-stage head and neck cancer, decreased SMC1B expression leads to potentially increased genome instability and greater cancer progression risk, which could account for the risk of SPT (Second Primary Tumor) and/or recurrence [80].
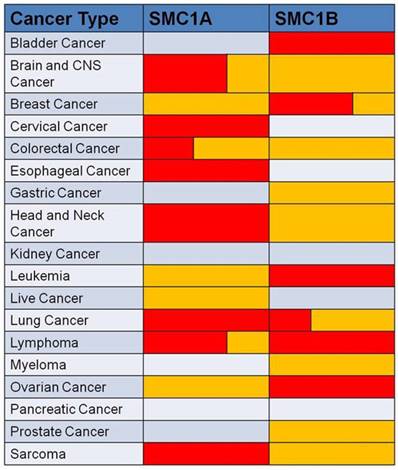
In conclusion, SMC1 expression is dysregulated in various human cancers associated with prognostic indicators. Aberrant expression of SMC1 is confirmed by analysis of SMC1 gene expression data setting at Oncomine (Figure 2).
Data showed SMC1 genes are aberrantly expressed in human cancers compared to their normal tissue. Red represents over-expression, and yellow represents low-expression in tumor versus normal tissue. The volume indicates the percentage of the two different expression forms.
SMC1 mutations are prevalent in human cancers
SMC1 mutations have been identified in human cancers. Being different from other cohesin mutations that were heterozygous in nature, SMC1A gene locates on chromosome X as a single allele, thereby its mutation cannot be heterozygous. The first somatic mutation of SMC1A in human tumors was reported in 2008 wherein Barber et al. identified somatic mutations in 4 out of 132 colorectal cancers (F396L/F, R434W, I560I/M and V1186I/V) [81]. Later, Cucco et al. screened 11 somatic mutations in early colorectal adenomas but only one mutation in colorectal carcinomas. SMC1A mutations decrease almost 5 fold during colon cancer development in early adenomas than in colorectal cancers. Mechanistically, overexpression of the mutations identified in early adenomas can cause chromosomal instability and aneuploidy [82].
In addition to colorectal cancer, there has been increasing evidences for SMC1 mutations in AML. In 2013, the Cancer Genome Atlas reported cohesin mutations in 26 of 200 de novo AMLs (13%) including 7 with SMC1A mutations (6/7 missense) [83]. Subsequently, the mutation frequency of SMC1A among 389 AML patients was evaluated by Thol et al., and found 2 male patients with 2 missense mutations in SMC1A (A078V, R816S) were identified [84]. Moreover, Kon et al. detected 4 different SMC1A mutations among 610 myeloid neoplasm samples (K190T, R586W, M689V and R807H). Interestingly, hemimethylation of the SMC1A promoter was found in two female cases. However, the expression of SMC1A was not significant different in 17 myeloid leukemia cell lines with or without known cohesin mutations. However, in cell lines (Kasumi-1, KG-1, P31FUJ, MOLM-7 and MOLM-13) with mutated or reduced expression of cohesin components, SMC1A is significantly reduced in the chromatin-bound fractions [85]. There is some evidence t that cohesin gene mutations may also act as an initiation of a subset of AMLs. Welch et al. indentified 2 SMC1A recurrent mutations in AML through whole genome sequencing [86]. In addition, a study by Yoshida et al. evaluated 2 SMC1A somatic mutations in DS-AMKL (Down Syndrome-related Acute Megakaryoblastic Leukemia) (R196H, E418_splice) [87]. Similarly, Huether et al. indentified 4 mutations of SMC1A in pediatric CBF AML (Core Binding Factor Acute Myeloid Leukemia) (*insR131, R92G, G1131R, R196C) [88].
In several other tumor types, mutations of SMC1A have been detected as well. Firstly, it was reported that SMC1A mutations were found in Ewing sarcoma, known as a very rare type of malignant tumor whose cancer cells were found in the pediatric bone and soft tissue. In the United States, about 200 children and young adults suffer from it annually and it shows a mild preference for males [89]. Substantial effort has been made to look for genes that are mutated in Ewing sarcoma tumors. Crompton et al. identified SMC1A mutations (1%, E141K) in addition to STAG2 mutations commonly in Ewing sarcoma [90]. In another large scale sequencing study, Guo et al. found occasional somatic mutations in SMC1A (3%) [91]. Then, 1 SMC1A mutation in pediatric Ependymoma (T182_F184del) was identified [88]. Finally, Balbas Martinez et al. identified 3% frequent somatic mutations in SMC1B in bladder cancer by exome sequencing (413, 188) [92]. Interestingly, transposon insertional mutagenesis in SMC1B mice were found to be more susceptible to intestinal tumor [93].
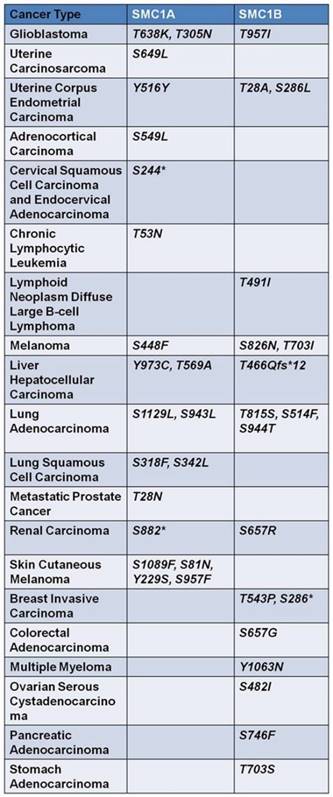
Taken together, these studies indicate that SMC1 mutation could be a significant event in several tumor types, albeit with a low mutation rate (1%-4%). Given that phosphorylation of SMC1 plays an important role in the maintenance of genome stability [66], mutations in the serine, threonine or tyrosine residues of SMC1 that can be phosphorylated have also been extensively studied, which were summarized in SMC1A and SMC1B data sets at cBioPortal (Figure 3).
Mutations of serine, threonine and tyrosine of the two isoforms of SMC1, widely existed in human cancers, are exhibited in the form above. These mutations dysfunction the effective phosphorylation of SMC1 proteins, thus interrupting maintenance of genome stability and DSB repair. Asterisk represents nonsense mutation.
Conclusion
As a core part of the cohesin complex, SMC1 is required for sister chromatid cohesion and maintenance of the genome integrity. Moreover, it can be activated to facilitate genome amplification under conditions of viral infection [8, 94, 95]. These characteristics make SMC1 a pivotal molecular linked to human tumorigenesis and ontogenesis [96].
We can surmise that SMC1 expression could be regulated by other unknown proteins or factors and its wax or wane interferes with genome stability which finally triggers the onset of cancer, from evidence that the expression quantity change of SMC1A and SMC1B was found to be existed widely in human cancers. Nonetheless, it is still elusive about pathways taken by SMC1 per se to complicate the maintenance of genome integrity or induced DNA damage checkpoint/repair through intricate protein interactions. Likewise, SMC1A mutations may lead to chromosomal instability by two different ways. On one hand, SMC1A mutations are directly associated with the classical functions of cohesion. These mutations might result in chromosome imbalance with chromosome loss or gain through influencing correct chromosome segregation. On the other hand, cohesion is recently found to be implicated in the regulation of gene expression. SMC1A mutations could contribute to tumorigenesis via regulating the expression of proto-oncogenes or tumor suppressor genes, given that cohesin is recently found to be implicated in the regulation of gene expression. Furthermore, the SMC1 activity can be regulated by phosphorylation and acetylation.
Here, we summarized the expression and phosphorylation mutations for the human SMC1 gene in cancers. In particular, we focus on the functional effects of SMC1 mutations as well as crucial questions that need to be addressed in future in order to provide a promising target for the treatment of malignant tumors.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China to Fei Yi (81502414); Ministry of Education Innovation Team Development Plan, Key Project of the National Science Foundation, National Program on Key Research Project to Liu Cao (IRT13101, 81130042, 31171323, 2016YFC1302400, 2015225003); National Science Fundation of China to Xiaoyu Song (31300963, 201302105) and Tingting Zhou (81502438); Science and Technology Research Project of Liaoning Provincial Education Department to Hongde Xu (LK201627); Natural Science Foundation of Liaoning Province of China to Ning Bai (20170541051).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986-1997
2. Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47-57
3. Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35-45
4. Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc Natl Acad Sci U S A. 2003;100:15387-15392
5. Yan R, McKee BD. The cohesion protein SOLO associates with SMC1 and is required for synapsis, recombination, homolog bias and cohesion and pairing of centromeres in Drosophila Meiosis. PLoS Genet. 2013;9:e1003637
6. Huber RG, Kulemzina I, Ang K. et al. Impairing Cohesin Smc1/3 Head Engagement Compensates for the Lack of Eco1 Function. Structure. 2016;24:1991-1999
7. Revenkova E, Eijpe M, Heyting C. et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555-562
8. Luo Y, Deng X, Cheng F. et al. SMC1-mediated intra-S-phase arrest facilitates bocavirus DNA replication. J Virol. 2013;87:4017-4032
9. Wakeman TP, Kim WJ, Callens S. et al. The ATM-SMC1 pathway is essential for activation of the chromium[VI]-induced S-phase checkpoint. Mutat Res. 2004;554:241-251
10. Cobbe N, Heck MM. Review: SMCs in the world of chromosome biology- from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123-143
11. Bauerschmidt C, Woodcock M, Stevens DL. et al. Cohesin phosphorylation and mobility of SMC1 at ionizing radiation-induced DNA double-strand breaks in human cells. Exp Cell Res. 2011;317:330-337
12. Schar P, Fasi M, Jessberger R. SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res. 2004;32:3921-3929
13. Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635-1648
14. Larionov VL, Karpova TS, Kouprina NY. et al. A mutant of Saccharomyces cerevisiae with impaired maintenance of centromeric plasmids. Curr Genet. 1985;10:15-20
15. Jessberger R, Frei C, Gasser SM. Chromosome dynamics: the SMC protein family. Curr Opin Genet Dev. 1998;8:254-259
16. Walker JE, Saraste M, Runswick MJ. et al. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945-951
17. Saitoh N, Goldberg IG, Wood ER. et al. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303-318
18. Stursberg S, Riwar B, Jessberger R. Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene. 1999;228:1-12
19. Beasley M, Xu H, Warren W. et al. Conserved disruptions in the predicted coiled-coil domains of eukaryotic SMC complexes: implications for structure and function. Genome Res. 2002;12:1201-1209
20. Camdere G, Guacci V, Stricklin J. et al. The ATPases of cohesin interface with regulators to modulate cohesin-mediated DNA tethering. Elife. 2015:4
21. Akhmedov AT, Frei C, Tsai-Pflugfelder M. et al. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088-24094
22. Kulemzina I, Ang K, Zhao X. et al. A Reversible Association between Smc Coiled Coils Is Regulated by Lysine Acetylation and Is Required for Cohesin Association with the DNA. Mol Cell. 2016;63:1044-1054
23. Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511-521
24. Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095-1101
25. Jessberger R, Chui G, Linn S. et al. Analysis of the mammalian recombination protein complex RC-1. Mutat Res. 1996;350:217-227
26. Jessberger R, Riwar B, Baechtold H. et al. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061-4068
27. Krishnan B, Thomas SE, Yan R. et al. Sisters unbound is required for meiotic centromeric cohesion in Drosophila melanogaster. Genetics. 2014;198:947-965
28. Haering CH, Lowe J, Hochwagen A. et al. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773-788
29. Anderson DE, Losada A, Erickson HP. et al. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419-424
30. Huis in 't Veld PJ, Herzog F, Ladurner R. et al. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968-972
31. Sumara I, Vorlaufer E, Gieffers C. et al. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749-762
32. Yeh C, Coyaud E, Bashkurov M. et al. The Deubiquitinase USP37 Regulates Chromosome Cohesion and Mitotic Progression. Curr Biol. 2015;25:2290-2299
33. Storre J, Schafer A, Reichert N. et al. Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J Biol Chem. 2005;280:41380-41386
34. Novak I, Wang H, Revenkova E. et al. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83-90
35. Adelfalk C, Janschek J, Revenkova E. et al. Cohesin SMC1beta protects telomeres in meiocytes. J Cell Biol. 2009;187:185-199
36. Biswas U, Wetzker C, Lange J. et al. Meiotic cohesin SMC1beta provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions. PLoS Genet. 2013;9:e1003985
37. Mannini L, Cucco F, Quarantotti V. et al. SMC1B is present in mammalian somatic cells and interacts with mitotic cohesin proteins. Sci Rep. 2015;5:18472
38. Losada A, Hirano T. Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays. 2001;23:924-935
39. Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269-1287
40. Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525-558
41. Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3-Scc1 gate. EMBO J. 2013;32:666-676
42. Guan J, Ekwurtzel E, Kvist U. et al. Cohesin protein SMC1 is a centrosomal protein. Biochem Biophys Res Commun. 2008;372:761-764
43. Gregson HC, Schmiesing JA, Kim JS. et al. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276:47575-47582
44. Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc Natl Acad Sci U S A. 2008;105:15441-15445
45. Gregson HC, Van Hooser AA, Ball AR Jr. et al. Localization of human SMC1 protein at kinetochores. Chromosome Res. 2002;10:267-277
46. Schmiesing JA, Ball AR Jr, Gregson HC. et al. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci U S A. 1998;95:12906-12911
47. Laugsch M, Seebach J, Schnittler H. et al. Imbalance of SMC1 and SMC3 cohesins causes specific and distinct effects. PLoS One. 2013;8:e65149
48. Kim JS, Krasieva TB, LaMorte V. et al. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149-45153
49. Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377-3388
50. Bauerschmidt C, Arrichiello C, Burdak-Rothkamm S. et al. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2010;38:477-487
51. Yazdi PT, Wang Y, Zhao S. et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571-582
52. Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560-570
53. Ivey RG, Moore HD, Voytovich UJ. et al. Blood-based detection of radiation exposure in humans based on novel phospho-Smc1 ELISA. Radiat Res. 2011;175:266-281
54. Kim BM, Choi JY, Kim YJ. et al. Reoxygenation following hypoxia activates DNA-damage checkpoint signaling pathways that suppress cell-cycle progression in cultured human lymphocytes. FEBS Lett. 2007;581:3005-3012
55. Helchowski CM, Skow LF, Roberts KH. et al. A small ubiquitin binding domain inhibits ubiquitin-dependent protein recruitment to DNA repair foci. Cell Cycle. 2013;12:3749-3758
56. Wakeman TP, Xu B. ATR regulates hexavalent chromium-induced S-phase checkpoint through phosphorylation of SMC1. Mutat Res. 2006;610:14-20
57. Musio A, Montagna C, Mariani T. et al. SMC1 involvement in fragile site expression. Hum Mol Genet. 2005;14:525-533
58. Musio A, Montagna C, Zambroni D. et al. Inhibition of BUB1 results in genomic instability and anchorage-independent growth of normal human fibroblasts. Cancer Res. 2003;63:2855-2863
59. So EY, Ausman M, Saeki T. et al. Phosphorylation of SMC1 by ATR is required for desferrioxamine (DFO)-induced apoptosis. Cell Death Dis. 2011;2:e128
60. Kitagawa R, Bakkenist CJ, McKinnon PJ. et al. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423-1438
61. Watrin E, Peters JM. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 2009;28:2625-2635
62. Li T, Wang ZQ. Point mutation at the Nbs1 Threonine 278 site does not affect mouse development, but compromises the Chk2 and Smc1 phosphorylation after DNA damage. Mech Ageing Dev. 2011;132:382-388
63. Antoccia A, Sakamoto S, Matsuura S. et al. NBS1 prevents chromatid-type aberrations through ATM-dependent interactions with SMC1. Radiat Res. 2008;170:345-352
64. Stewart GS, Wang B, Bignell CR. et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961-966
65. Wu J, Prindle MJ, Dressler GR. et al. PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J Biol Chem. 2009;284:18078-18084
66. Diaz-Martinez LA, Beauchene NA, Furniss K. et al. Cohesin is needed for bipolar mitosis in human cells. Cell Cycle. 2010;9:1764-1773
67. Wong RW. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle. 2010;9:198-200
68. Hodges CA, Revenkova E, Jessberger R. et al. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet. 2005;37:1351-1355
69. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674
70. Wang J, Yu S, Cui L. et al. Role of SMC1A overexpression as a predictor of poor prognosis in late stage colorectal cancer. BMC Cancer. 2015;15:90
71. Li J, Feng W, Chen L. et al. Downregulation of SMC1A inhibits growth and increases apoptosis and chemosensitivity of colorectal cancer cells. J Int Med Res. 2016;44:67-74
72. Yadav S, Sehrawat A, Eroglu Z. et al. Role of SMC1 in overcoming drug resistance in triple negative breast cancer. PLoS One. 2013;8:e64338
73. Li K, Ying M, Feng D. et al. SMC1 promotes epithelial-mesenchymal transition in triple-negative breast cancer through upregulating Brachyury. Oncol Rep. 2016;35:2405-2412
74. Ma Z, Lin M, Li K. et al. Knocking down SMC1A inhibits growth and leads to G2/M arrest in human glioma cells. Int J Clin Exp Pathol. 2013;6:862-869
75. Yang Y, Zhang Z, Wang R. et al. siRNA-mediated knockdown of SMC1A expression suppresses the proliferation of glioblastoma cells. Mol Cell Biochem. 2013;381:209-215
76. Ansari D, Andersson R, Bauden MP. et al. Protein deep sequencing applied to biobank samples from patients with pancreatic cancer. J Cancer Res Clin Oncol. 2015;141:369-380
77. Hsiao YH, Su YA, Tsai HD. et al. Increased invasiveness and aggressiveness in breast epithelia with cytoplasmic p63 expression. Int J Biol Sci. 2010;6:428-442
78. Kim KH, Yoo HY, Joo KM. et al. Time-course analysis of DNA damage response-related genes after in vitro radiation in H460 and H1229 lung cancer cell lines. Exp Mol Med. 2011;43:419-426
79. Homme C, Krug U, Tidow N. et al. Low SMC1A protein expression predicts poor survival in acute myeloid leukemia. Oncol Rep. 2010;24:47-56
80. Zhang X, Yang H, Lee JJ. et al. MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis. 2010;31:2118-2123
81. Barber TD, McManus K, Yuen KW. et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443-3448
82. Cucco F, Servadio A, Gatti V. et al. Mutant cohesin drives chromosomal instability in early colorectal adenomas. Hum Mol Genet. 2014;23:6773-6778
83. Ley TJ, Miller C, Ding L. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059-2074
84. Thol F, Bollin R, Gehlhaar M. et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123:914-920
85. Kon A, Shih LY, Minamino M. et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232-1237
86. Welch JS, Ley TJ, Link DC. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264-278
87. Yoshida K, Toki T, Okuno Y. et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293-1299
88. Huether R, Dong L, Chen X. et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630
89. Esiashvili N, Goodman M, Marcus RB Jr. Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. 2008;30:425-430
90. Crompton BD, Stewart C, Taylor-Weiner A. et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326-1341
91. Cha EK, Bochner BH. Re: whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Eur Urol. 2015;67:350-351
92. Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E. et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464-1469
93. March HN, Rust AG, Wright NA. et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202-1209
94. Mehta K, Gunasekharan V, Satsuka A. et al. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog. 2015;11:e1004763
95. Anacker DC, Gautam D, Gillespie KA. et al. Productive replication of human papillomavirus 31 requires DNA repair factor Nbs1. J Virol. 2014;88:8528-8544
96. Strunnikov A. Cohesin complexes with a potential to link mammalian meiosis to cancer. Cell Regen (Lond). 2013;2:4
Author contact
![]() Corresponding author: xysongedu.cn
Corresponding author: xysongedu.cn

 Global reach, higher impact
Global reach, higher impact