ISSN: 1449-2288
Int J Biol Sci 2018; 14(3):306-320. doi:10.7150/ijbs.23869 This issue Cite
Research Paper
Environmental adaptation of Acanthamoeba castellanii and Entamoeba histolytica at genome level as seen by comparative genomic analysis
1. Institute of Bioinformatics, University Münster, Niels-Stensen Strasse 14, Münster 48149, Germany
2. Laboratory of Bioenergetics, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University
3. Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba 277-8562, Japan
Abstract
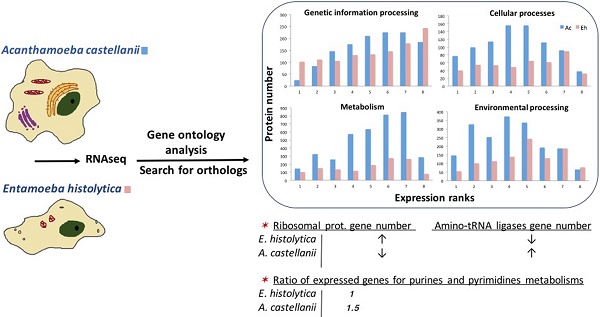
Amoebozoans are in many aspects interesting research objects, as they combine features of single-cell organisms with complex signaling and defense systems, comparable to multicellular organisms. Acanthamoeba castellanii is a cosmopolitan species and developed diverged feeding abilities and strong anti-bacterial resistance; Entamoeba histolytica is a parasitic amoeba, who underwent massive gene loss and its genome is almost twice smaller than that of A. castellanii. Nevertheless, both species prosper, demonstrating fitness to their specific environments. Here we compare transcriptomes of A. castellanii and E. histolytica with application of orthologs' search and gene ontology to learn how different life strategies influence genome evolution and restructuring of physiology. A. castellanii demonstrates great metabolic activity and plasticity, while E. histolytica reveals several interesting features in its translational machinery, cytoskeleton, antioxidant protection, and nutritional behavior. In addition, we suggest new features in E. histolytica physiology that may explain its successful colonization of human colon and may facilitate medical research.
Keywords: Life strategy, genomic adaptation, comparative genomics, Acanthamoeba castellanii, Entamoeba histolytica

 Global reach, higher impact
Global reach, higher impact