10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(7):726-735. doi:10.7150/ijbs.24505 This issue Cite
Research Paper
FBW7 suppresses metastasis of colorectal cancer by inhibiting HIF1α/CEACAM5 functional axis
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, China;
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China;
3. Departments of CyberKnife, Huashan Hospital, Fudan University, Shanghai 200032, China.
*Qingguo Li and Yaqi Li contributed equally to this work.
Received 2017-12-22; Accepted 2018-4-10; Published 2018-5-12
Abstract
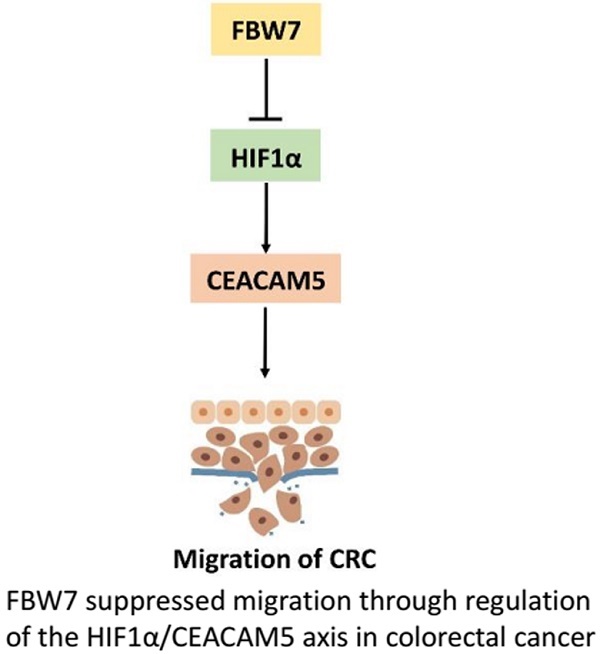
F-box and WD repeat domain-containing 7 (FBW7) functions as a major tumor suppressor by targeting oncoproteins for degradations. FBW7 has been reported to be one of the most frequently mutated genes in colorectal cancer (CRC). However, its roles and possible mechanisms in the development of CRC are still unclear. In the present study, we adopted immunohistochemistry staining in tissue microarray (TMA), consisting of 276 samples from stage I-IV CRC patients, and analyzed the correlation between FBW7 expression and clinicopathological parameters, as well as overall survival (OS) and disease-free survival (DFS). The impact of FBW7 on migration was further validated in vitro. Whole-genome expression microarray (GEO,accession numbers GSE76443), was then analyzed to find the possible target of FBW7. The results were verified by functional experiments in vitro and IHC staining of TMA. Finally, luciferase and chromatin immunoprecipitation (ChIP) assays were carried out to identify the possible mechanisms. The expression level of FBW7 in TMA was negatively correlated with serum CEA level, venous invasion, N stage and M stage, and positively associated with the survival of CRC patients(P<0.05). Ectopic FBW7 expression significantly suppressed migration of colon cancer cells in vitro. GEO analysis revealed that decreased FBW7 significantly correlated with increased level of CEACAM5, which encoded CEA. The correlation was verified by IHC of TMA and silencing CEACAM5 inhibited migration in vitro. Mechanistically, we demonstrated that CEACAM5 was a HIF1α target gene and that FBW7 regulated CEACAM5 in a HIF1α-dependent manner. In conclusion, our results revealed that FBW7 suppressed migration through regulation of the HIF1α/CEACAM5 axis in colorectal cancer. Therefore, our study sheds novel lights on the impact of FBW7 on HIF1α/CEACAM5 signaling axis and constitutes potential prognostic predictors and therapeutic targets for CRC.
Keywords: FBW7, Colorectal cancer, HIF1α, CEACAM5
Introduction
Colorectal cancer (CRC) is the third frequently diagnosed cancer and is also the third leading cause of cancer death worldwide[1]. Despite the treatments have been improved dramatically in recent years, invasion and metastasis, the major causes of CRC related relapse and death, greatly impeded the treatment efficiency. About 20-45% of patients who underwent curative resection developed recurrence or metastasis[2, 3]. However, the molecular mechanisms underlying the invasion and migration of CRC are still limited.
FBW7 (F-box and WD repeat domain-containing 7), also known as Fbxw7, has been found to be involved in numerous cellular processes including cell proliferation, apoptosis, cell cycle and differentiation [4-6]. Loss of the tumor suppressive function of FBW7 has been proposed to drive the progression of multiple cancers, including gastric cancer, pancreatic cancer, melanoma, and breast cancer[7-10]. FBW7 belongs to one of F-box proteins and is an essential component of SCF (SKP1-CUL1-F-box) E3 ligase complex, which targets proteins for ubiquitination and degradation[9, 11]. In recent years, there are some FBW7 E3 ligase substrates identified, including c-Myc, HIF1α, Notch1, KLF5, Cyclin E1, and Mcl-1[4, 12]. Loss of FBW7 in cancer leads to aberrant accumulation of these substrates, accounting for the tumor phenotypes observed in xenograft or genetic mouse models[9, 12].
FBW7 is one of the eight most frequently mutated genes in non-hypermutated CRC with its mutation rate of approximately 11%[13]. FBW7 pathway mutations cause genomic instability and induce p53 activity. It has been reported that FBW7 and p53 synergistically suppress adenocarcinomas that mimic advanced human CRC with respect to histopathology, metastasis, and chromosomal instability[14]. FBW7 mediates drug resistance of CRC to targeted therapies by blocking Mcl-1 degradation [15]. Recent studies suggest that FBW7 may have functions in cell migration and metastasis. It is reported that FBW7 suppresses the ENO1-induced gene expression, lactate production, cell proliferation and migration in CRC[16]. However, as a general tumor suppressor in CRC, the expression of FBW7 and its exact function in CRC progression and metastasis are unknown.
To investigate the role of FBW7 in CRC progression, we used tissue microarray (TMA) and immunohistochemistry (IHC) to examine the expression of FBW7 protein in the different stages of human CRC lesions. Our data indicated that FBW7 staining was significantly reduced in primary tumor compared with normal control. Notably, we observed that the increased expression of FBW7 was significantly correlated with a better 5-year overall survival (OS) and disease free survival (DFS). Our in vitro study showed significant inhibition of colon cancer cell migration and invasion by ectopic FBW7 expression. Mechanically, FBW7 negatively regulates colon cancer invasion by targeting the HIF1α/ CEACAM5 axis.
Methods
Patient specimens and TMA construction
We recruited 276 formalin-fixed, paraffin-embedded CRC tissues from between January 2007 and November 2009 archives of the Department of Pathology at Fudan University Shanghai Cancer Center (FUSCC). Construction of this TMA has been previously described in detail[17]. Patients included in the case cohort were under intact follow-up. Signed informed consent for the research proposes of clinical samples was obtained from every patient. The clinicopathological variables were collected from patients' medical records, and disease clinical stages were classified according to the 8th UICC/AJCC CRC TNM staging system.
Immunohistochemical staining
TMA were deparaffinized, rehydrated, and then incubated with rabbit polyclonal antibody against FBW7 (A301-720A, Bethyl), CEACAM5 (10421-1-AP, Proteintech), HIF1α (20960-1-AP, Proteintech) at 4°C overnight following heat-induced epitope retrieval. Staining detection was performed using the GTVisionTM III Kit (GK500705, Gene Tech, Shanghai, China) detection kit according to the manufacturer's instructions. PBS was used as negative control throughout. A semi-quantitative scoring system was used[18], which evaluated both staining intensity (0, no stain; 1+, weak stain; 2+, moderate stain; 3+, strong stain) and the percentage of stained cells (0,<5%; 1, 5-25%; 2, 26-50%; 3, 51-75%; and 4, >75%). Scores for staining intensity and percentage positivity of cells were then multiplied to generate the immunoreactivity score (IS) for each case. All cases were sorted into two groups according to the IS. High expression of FBW7 and HIF1α were defined as detectable immunoreactions in nucleus while CEACAM5 was stained in membranes with IS≥4[19].
Cell culture and reagents
Colon cancer cell line HCT116, LoVo, RKO, SW480, and SW620 were originally obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The cells were cultured in medium according to The Defense Technical information Center recommendation supplemented with 10% FBS (Gibco, Life Technology, Austria), 1% penicillin/ streptomycin in a humidified 5% (v/v) atmosphere of CO2 at 37℃.
Plasmids, siRNA and transfection
The Flag-tagged coding sequence of human FBW7 was cloned into the lentiviral vector pCDH-CMV-MCS-EF1-puro (SBI, USA) to generate FBW7 expression plasmids. The siRNA of CEACAM5 targeting sequences were: 5'- GACCCUCACUCUAUUCAAU-3', 5'- CAGUACUCUUGGUUUGUCA-3' and 5'-CAAGCCCAUAACUCAGACA-3'. Cells were seeded at an approximate concentration and were cultured under standard incubation conditions for 24 hours before transfection. FBW7 expression and control vectors, and CEACAM5 and control siRNAs were transfected using Lipofectamine 2000 (Invitrogen).
Cell proliferation assay
Cells were seeded and cultured in 96-well plates. A CCK8 assay (Dojindo) was performed, and the optical density (OD) at 450 nm was measured in an automatic microplate reader (BioTek). Each experiment was performed in triplicate and repeated at least twice.
Cell migration assay
Cell migration ability was analyzed by the Transwell chamber assay. Cells were plated in medium without serum, and medium containing 10% FBS in the lower chamber served as chemoattractant. After 48 hours of incubation, the cells that did not migrate or invade through the pores were carefully wiped out with cotton wool. Then the inserts were stained with 20% methanol and 0.2% crystal violet, imaged, and counted with an inverted microscope.
RNA isolation and quantitative real-time PCR
Total RNA was prepared using TRIzol reagent (Invitrogen), and cDNA was obtained by reverse transcription using a TaKaRa PrimeScript RT reagent Kit (RR036A). The expression status of candidate genes and GAPDH were determined by quantitative real-time PCR using an ABI 7900HT Real-Time PCR system (Applied Biosystems). Primers used were human FBW7: 5'-CCACTGGGCTTGTACCATGTT-3' (forward), and 5'-CAGATGTAATTCGGCGTCGTT-3' (reverse); human GAPDH: 5'-CGACCACTTTGTCAAGCTCA-3' (forward), and 5'-AGGGGAGATTCAGTGTGGTG-3' (reverse). All reactions were run in triplicate.
Western blot analysis
Equal amounts of cell lysates were subjected to 10% SDS-PAGE, and proteins were transferred onto PVDF membranes (Biorad). The membranes were probed overnight with specific primary antibodies, which were detected with corresponding secondary antibodies (Cell Signaling Technology). The immunoreactive bands were visualized using enhanced chemiluminescence (Thermo Scientific). The following primary antibodies were used: FBW7 (A301-721A, Bethyl), CEACAM5 (10421-1-AP, Proteintech), HIF1α (20960-1-AP, Proteintech) and GAPDH (10494-1-AP, Proteintech). GAPDH served as a loading control.
Luciferase assays
For the luciferase assays, the CEACAM5 promotor was cloned into the pGL3 basic vector (Promega). Then HCT-116 and LoVo cells (8×104 cells/well) were cultured in 96-well plates and co-transfected with the pGL3-CEACAM5 or pGL3-control, pCDH-FBW7 or pCDH-control and Renilla plasmid using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were lysed using 20μL of passive lysis buffer. Next, a dual-luciferase assay was carried out as described by the manufacturer (Promega). The ratio of firefly to Renilla luciferase activity was used to express luciferase activities. All experiments were performed in triplicate.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-ChIP Kit from Millipore according to the manufacturer's protocol. Primers to detect CEACAM5 promoter occupancy were: F: 5′-AAGACAGGTTGTCCTCCCAGGGGA-3′ and R: 5′- CCTCCTCTGTGGAGAAGAGCTTGA -3′.
Statistical analysis
All data were shown as mean ± SD. The Student t test was used for statistical analysis unless otherwise noted, with P < 0.05 considered significant. In IHC assays of CRC samples, Spearman's Rank correlation assay was used to determine the correlation between FBW7 and clinicopathological factors. Survival analysis was performed using the Kaplan-Meier and log-rank method. All data were analyzed using SPSS 21.0 statistical package (SPSS, Chicago, IL).
Results
High FBW7 staining correlates with better survival outcomes of CRC
In total, 276 patients with CRC were included in TMA with age ranging from 27 to 85 years (median 58). There were 166 (60.1%) male and 110 (39.9%) female cases. There were 24 (8.7%) tumors at stage I, 90 (32.6%) at stage II, 120 (43.5%) at stage III, and 42 (15.2%) at stage IV. Table 1 summarized the baseline characteristics of patients in TMA.
IHC staining was performed to study the expression level of FBW7 on TMA. The FBW7 staining was predominantly detected in the nucleus as demonstrated in Fig 1a. Notably, FBW7 staining was significantly reduced as the tumor advanced (Fig.1b). Specifically, FBW7 is negatively correlated with CEA levels (P=0.037) and venous invasion (P=0.029) (Table 1). But we did not observe any significant correlation between nuclear FBW7 staining and other clinicopathological factors. These suggested that FBW7 may have a role in CRC progression and metastasis.
Overexpression of FBW7 correlates with better survival of patients with colorectal cancer. (a) Immunohistochemical (IHC) staining of FBW7 was performed TMA of 276 patients with colorectal cancer. The FBW7 staining was predominantly in the nucleus. The staining of FBW7 in normal tissue was shown in picture ① and ②, and that in tumor tissue was shown in picture ③ and ④(scale bar, 50μm).(b) FBW7 staining was significantly reduced as the pathological stage of the tumor advanced. (c) Patients with colorectal cancer were then stratified into FBW7-low and FBW7-high subgroups. Kaplan-Meier curves showed that 5-year overall survival (OS) was lower of patients in FBW7-low subgroup than those in FBW7-high subgroup (36.8% vs. 78.3%, χ2=47.415, P<0.001). (d) Then disease-free survival was analyzed and forty-two patients with distant metastases were excluded from the study. Kaplan-Meier curves indicated that the 5-year DFS were 44.7% and 80.0% for patients with low and high FBW7 expression (χ2=32.769, P<0.001).
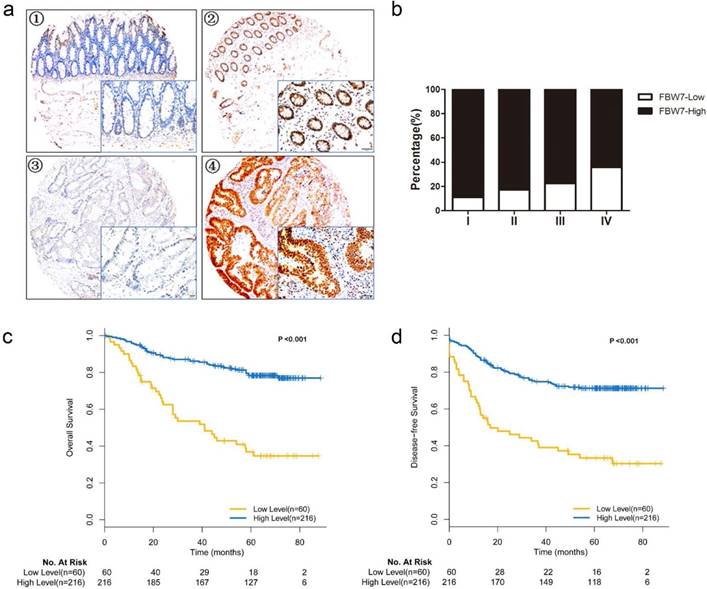
To evaluate the association between FBW7 expression and survival outcomes among patients with CRC, we stratified the patients into FBW7-low and FBW7-high expression subgroups and then used Kaplan-Meier curves to compare the overall survival (OS) and disease-free survival (DFS) of the two subgroups (Fig.1c, d). The analysis showed that the rate of 5-year OS was lower among the patients with FBW7-low tumors than among those with FBW7-high expression tumors (36.8% vs. 78.3%, χ2=47.415, P<0.001). When analyzing the DFS, forty-two patients with distant metastases were excluded from the study, the results indicated that the 5-year DFS were 44.7% and 80.0% for those with low and high FBW7 expression (χ2=32.769, P<0.001). Furthermore, multivariate Cox regression analysis showed that FBW7 expression was an independent factor for predicting both OS (HR: 0.280, 95%CI 0.175-0.448, P<0.001) and DFS (HR 0.216, 95%CI 0.123-0.378, P<0.001) (Table 2).
Inverse correlation between FBW7 levels and cell migration capacity in vitro
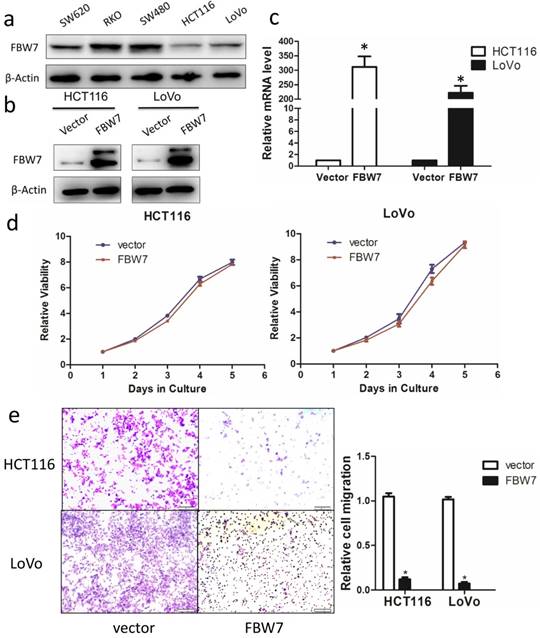
Having shown that decreased FBW7 expression associated with advanced tumor stage, we hypothesized that FBW7 might have an effect on the migration of colon cancer cells. We ectopically overexpressed FBW7 in HCT116 and LoVo cells, which exhibited the lowest endogenous FBW7 expression (Fig.2a). The overexpression efficiency was determined by Western blotting (Fig.2b) and qRT-PCR (Fig.2c). First, proliferation was examined using the CCK-8 assay and we found that overexpression of FBW7 did not impaired cell growth ability. (Fig.2d). Next, we used Transwell assay to investigate whether forced FBW7 expression had an effect on cell migration. As shown in Fig.2e, the number of cells having migrated through the filter was reduced up to four-fold in FBW7 overexpressing cells, indicating that FBW7 inhibits cell migration.
Overexpression of FBW7 inhibits the migration capacity of colon cells. (a) The baseline expression of FBW7 in different colon cancer cell lines was firstly detected by Western blotting and HCT 116 and LoVo cells showed the lowest expression of FBW7. We then ectopically overexpressed FBW7 in HCT 116 and LoVo cells, and the overexpression efficacy was determined by Western blotting (b) and qRT-PCR (c). (d) Overexpression of FBW7 did not change the proliferation capacity of HCT-116 and LoVo cells, which was examined by CCK-8 assay. (e) Overexpression of FBW7 inhibited the migration capacity of HCT-116 and LoVo cells, which was detected by Transwell migration assay.*P<0.05.
CEACAM5 is a target of FBW7 in colorectal cancer
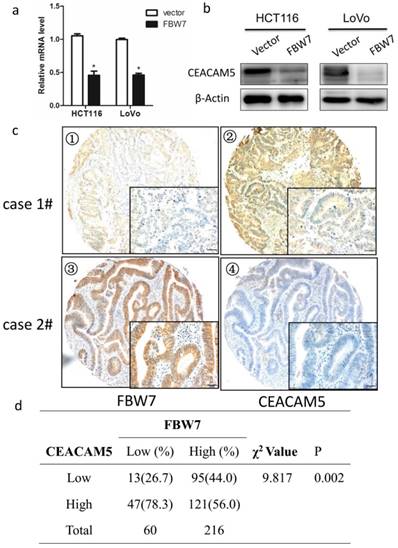
To identify the possible molecular mechanism in colon cancer in response to FBW7 overexpression, we studied a whole-genome expression microarray (GEO: accession numbers GSE76443), and found decreased FBW7 expression significantly correlated with increased level of CEACAM5, which encoded CEA that was widely used clinically as both a blood and tissue tumor marker of epithelial malignancy, especially for CRC. Besides, as mentioned above, FBW7 expression of TMA were inversely associated with serum CEA levels (Table 1). Hence, the results suggest that CEACAM5 is a target of FBW7. To test our hypothesis, we found ectopic FBW7 expression in HCT116 and LoVo cells significantly downregulated CEACAM5 on both protein and transcriptional level (Fig.3a and 3b). Next, we examined the expression of FBW7 and CEACAM5 in TMA by IHC staining and observed a negative correlation (χ2=9.817, P=0.002) (Fig. 3c and 3d). Together, these data strongly indicates that FBW7 negatively regulates CEACAM5 in CRC.
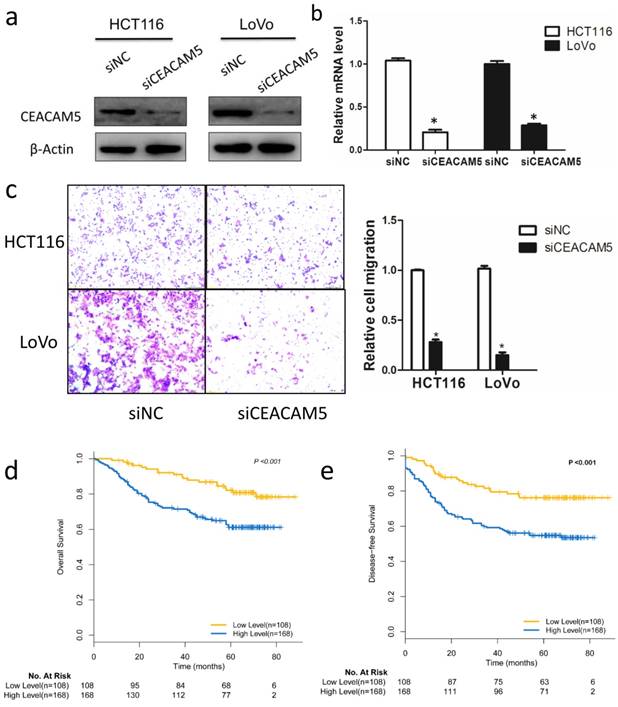
CEACAM5 contributes to colon cancer migration
Although CEA has been widely used as a prognostic marker in CRC, the functional role of CEACAM5 in vitro, especially on colon cancer cells migration, is not fully studied. To characterize the biological roles of CEACAM5, siRNA targeting CEACAM5 was employed in HCT116 and LoVo cell lines. Western blot and qRT-PCR analysis confirmed the knockdown efficiency 48h after siRNA transfection (Fig. 4a, 4b). A subsequent analysis indicated that silencing FBW7 inhibited HCT116 and LoVo cell migration by Transwell analysis (Fig. 4c). Then, we also found that high expression of CEACAM5 in TMA was significantly correlated with a poorer OS (χ2 =19.696, P<0.001) (Fig. 4d) and DFS (χ2 =10.687, P=0.001) (Fig. 4e), which is consistent with the previously published results of serum CEA in CRC.
Association between FBW7 expression and clinicpathological factors in colon cancer TMA (n = 276)
| Variable | n | FBW7 Expression | χ2 Value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Gender | 0.754 | 0.385 | |||
| Male | 166 | 39(65.0) | 127(58.8) | ||
| Female | 110 | 21(35.0) | 89(41.2) | ||
| Age | 0.082 | 0.774 | |||
| ≤60 | 170 | 36(60.0) | 134(62.0) | ||
| >60 | 106 | 24(42.0) | 82(38.0) | ||
| Primary site | 0.078 | 0.780 | |||
| Colon | 129 | 29(48.3) | 100(46.3) | ||
| Rectum | 147 | 31(51.7) | 116(53.7) | ||
| Histological type | 1.253 | 0.263 | |||
| Adenocarcinoma | 261 | 93(95.9) | 168(93.9) | ||
| Mucinous/SRCC* | 15 | 4(4.1) | 11(6.1) | ||
| T category | 3.493 | 0.174 | |||
| T1/T2 | 43 | 5(8.3) | 38(17.6) | ||
| T3 | 54 | 11(18.3) | 43(19.9) | ||
| T4 | 179 | 44(73.3) | 135(62.5) | ||
| N stage | 6.814 | 0.033 | |||
| N0 | 120 | 21(35.0) | 99(45.8) | ||
| N1 | 83 | 15(25.0) | 67(31.0) | ||
| N2 | 73 | 24(40.0) | 50(23.1) | ||
| M stage | 5.687 | 0.017 | |||
| M0 | 234 | 45(75.0) | 189(87.5) | ||
| M1 | 15 | 23(25.0) | 27(12.5) | ||
| Pathological grading | 3.535 | 0.316 | |||
| High | 59 | 17(28.3) | 42(19.4) | ||
| Moderate | 185 | 35(58.3) | 150(69.4) | ||
| Poor/ undifferentiation | 16 | 5(8.3) | 11(5.1) | ||
| Unknown | 16 | 3(5.0) | 13(6.0) | ||
| Venous invasion | 7.074 | 0.029 | |||
| Negative | 188 | 34(56.6) | 154(71.3) | ||
| Positive | 88 | 26(43.4) | 62(28.7) | ||
| Perineural invasion | 0.496 | 0.481 | |||
| Negative | 231 | 52(86.7) | 179(82.9) | ||
| Positive | 45 | 8(13.3) | 37(17.1) | ||
| CEA (μl/ml) | 6.582 | 0.037 | |||
| ≤5 | 170 | 29(48.3) | 141(65.3) | ||
| >5 | 95 | 29(48.3) | 66(30.6) | ||
| Unknown | 11 | 2(3.3) | 9(4.2) | ||
*SRCC: signet-ring cell carcinoma
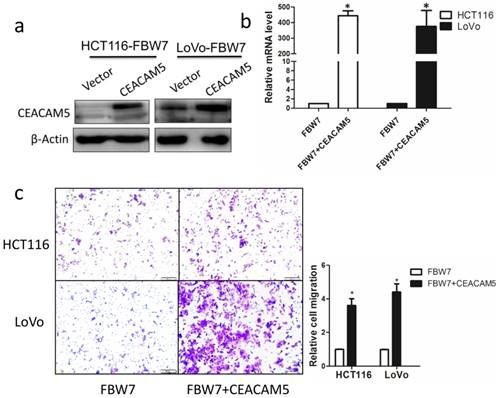
Overexpression of CEACAM5 reverses the inhibitory effects of FBW7 on migration in colon cancer cells
To further determine whether the function of FBW7 in CRC is mediated by CEACAM5, we restored CEACAM5 expression in FBW7-transfected HCT116 and LoVo cells. The restore effect was determined by Western blot and qRT-PCR (Fig. 5a, and b). Transwell migration assay showed that overexpression of CEACAM5 fully abolished the suppression effect of cell migration induced by FBW7 in both HCT116 and LoVo cells (Fig. 5c). These results further confirm that CEACAM5 is regulated by FBW7. FBW7 down-regulation may participate in CRC progression through potentiation of CEACAM5 expression.
Multivariate Cox proportional hazards analysis of Fbw7 expression and overall survival and disease free survival for patients with colorectal cancer.
| Overall survival | Disease free survival | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P | HR (95% CI) | P |
| Gender | 0.830(0.514-1.132) | 0.448 | 0.818(0.475-1.407) | 0.468 |
| Age | 1.238(0.764-2.005) | 0.387 | 1.143(0.674-1.937) | 0.619 |
| Primary site | 0.764(0.482-1.213) | 0.254 | 1.037(0.674-1.937) | 0.892 |
| Histological type | 0.779(0.246-2.468) | 0.671 | 0.967(0.316-2.962) | 0.953 |
| T category | 1.786(1.092-2.921) | 0.021 | 1.669(1.065-2.709) | 0.026 |
| N stage | 1.504 (1.098-2.061) | 0.011 | 1.799(1.301-2.488) | <0.001 |
| M stage | 5.935(3.414-10.319) | <0.001 | NI* | |
| Grade | 1.333(0.893-1.990) | 0.160 | 1.178(0.768-1.809) | 0.453 |
| Venous invasion | 1.113(0.694-1.784) | 0.658 | 1.217(0.725-2.044) | 0.458 |
| Perineural invasion | 1.308(0.740-2.312) | 0.356 | 2.001(1.042-3.854) | 0.037 |
| CEA | 0.716(0.454-1.129) | 0.151 | 1.046(0.610-1.794) | 0.869 |
| Fbw7 | 0.280(0.175-0.448) | <0.001 | 0.216(0.123-0.378) | <0.001 |
*Stage IV patients were excluded from disease free survival.
Abbreviation: CI, confidence interval; HR, hazard ratio.
Bold type indicates statistical significance.
FBW7 negatively regulates CEACAM5 in colorectal cancer. Overexpression of FBW7 downregulated CEACAM5 in both HCT116 and LoVo cells, which was determined by both qRT-PCR (a) and Western blotting (b). (c) Representative figures of the correlation of FBW7 and CEACAM5 were evaluated by IHC staining of TMA (scale bar, 50μm). (d) A negative correlation of FBW7 and CEACAM5 was validated (χ2=9.817, P=0.002). *P<0.05.
CEACAM5 contributes to colon cancer migration. The functional role of CEACEM5 in vitro was then examined in colon cancer cells. HCT-116 and LoVo cells were transfected with siRNA targeting CEACAM5, and the knockdown efficacy was validated by Western blotting (a) and qRT-PCR (b). Transwell migration assay indicated that knockdown of FBW7 inhibited migration capacity of HCT-116 and LoVo cells (c). Kaplan-Meier curves according to IHC results of CEACAM5 in TMA showed that 5-year OS (d) and DFS (e) was lower for patients in CEACAM5-high subgroup compared with those in CEACAM5-low subgroup. *P<0.05.
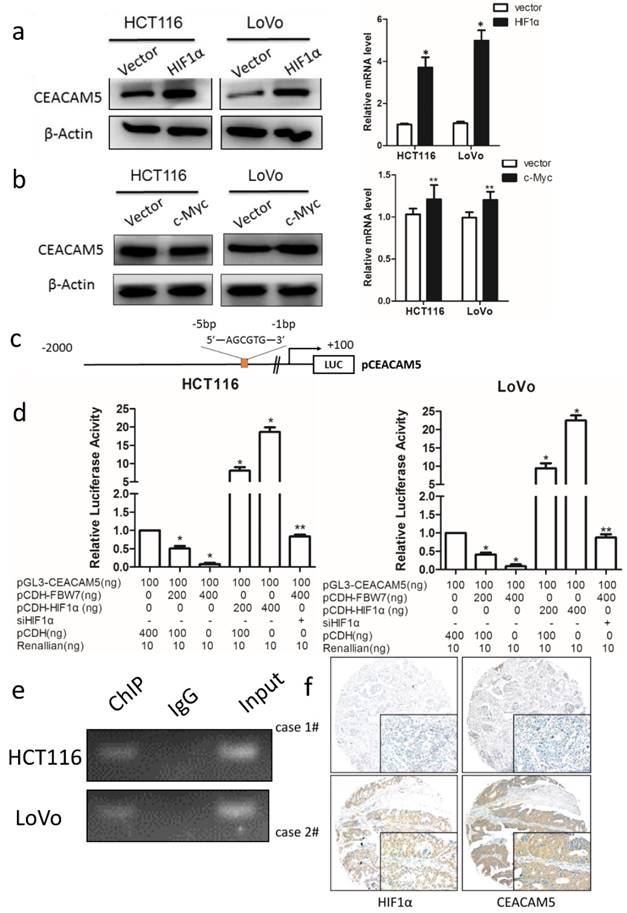
FBW7 regulates CEACAM5 expression in a HIF1α-dependent manner
FBW7 is an E3 ubiquitin ligase and targets many substrates for proteasomal degradation. Among these substrates, HIF1α, and c-Myc are well-known regulators of metabolism. Therefore, we first transfected HIF1α and c-Myc, respectively, into HCT116 and LoVo cells and found HIF1α could promote CEACAM5 expression on both transcriptional and protein levels (Fig 6a). However, no change in the expression of CEACAM5 was observed upon c-Myc transfected cell (Fig 6b). To further determine whether FBW7 regulates CEACAM5 through HIF1α, we analyzed the promoter region of CEACAM5 and found it harbored one E-box elements AGCGTG from -1 to -5bp (Fig.6c). We then cloned the promoter region of CEACAM5 from +100 to -2000bp into the pGL3-basic vector and performed a dual luciferase assay to investigate whether HIF1α and FBW7 influenced CEACAM5 promoter activity. As observed, HIF1α and FBW7 altered CEACAM5 promoter activity in a dosage dependent way (Fig. 6c). When co-transfected with siRNA against HIF1α and FBW7, FBW7 lost its inhibition effect on CEA promoter (Fig. 6d). Moreover, HIF1α occupied the E-boxes in the CEACAM5 promoter region, as determined by chromatin immunoprecipitation (ChIP) assay (Fig. 6e). In supporting the clues obtained from in vitro experiments with cell lines, we analyzed the expressional correlation in TMA between HIF1α and CEACAM5 levels in CRC samples. We observed that HIF1α level strongly correlated with CEACAM5 level in CRC tissue samples, and serum CEA levels were significantly higher in HIF1α-high patients (Fig. 6f). These findings suggest that FBW7 regulates CEACAM5 expression in a HIF1α-dependent manner. Fig.7. summarized the model of the mechanism of FBW7-mediated regulation of migration via the HIF1α/CEACAM5 axis in colorectal cancer.
Discussion
In this study, we found decreased FBW7 expression in CRC tissues compared with adjacent normal colorectal tissues. There was significantly inverse correlation between FBW7 overexpression and advanced tumor stage. CRC at higher stage possessed high invasive abilities and are more likely to spread to other areas of the body than early CRC. Thus, FBW7 might play an important role in malignant progression of CRC. Functional study indicated that FBW7 could inhibit the migration of the colon cancer cells. Regarding mechanisms, we found CEACAM5 might be a target of FBW7 from a public available whole-genome expression microarray, and we validated it with in vitro evidence. FBW7 is not a transcriptional factor. It functions as a substrate recognition and binding component in SCF (Skp1-Cul1-F-box protein-Rbx1) ubiquitin ligase complex and mediates the degradation of several important proteins[5]. We further identified HIF1α as a substrate that involved in FBW7 regulating CEACAM5 expression, and HIF1α also was confirmed as a transcriptional factor regulating CEACAM5 expression.
Overexpression of CEACAM5 reverses the inhibitory effects of FBW7 on migration in colon cancer cells. CEACAM expression was restored in FBW7-transfected HCT-116 and LoVo cells and the restore efficacy was determined by Western blotting (a) and qRT-PCR (b). Transwell migration assay indicated that overexpression of CEACAM5 fully abolished suppression effect on cell migration of FBW7 in both HCT-116 and LoVo cells (c). *P<0.05.
As a well-known tumor suppressor, FBW7 expression was significantly reduced in gastric cancer, CRC, pancreatic cancer, melanoma, and glioma[8, 20-23]. However, reports concerning FBW7 protein in CRC are limited. Iwatsuki et al. reported that patients with low FBW7 mRNA expression had significantly poorer prognosis. They found silencing FBW7 upregulated c-MYC and cyclin E, and promoted cell proliferation in vitro[22], but they did not investigate the prognostic value of FBW7 in CRC and neither the function of FBW7 in CRC migration and progression. Zhan et al. found that FBW7 inhibited glycolysis by physically binding to ENO1 and targets ENO1 for ubiquitin-mediated degradation [16]. Tong et al. demonstrated FBW7 mutations mediated drug resistance of CRC to targeted therapies by blocking Mcl-1 degradation[15]. Our studies revealed a new mechanism that FBW7 involved in tumor progression and provided novel predictive and treatment targets for CRC.
CEA, encoded by CEACAM5, was firstly identified as a tumor associated antigen for CRC. Now, it is clearly established that patients with elevated preoperative CEA levels have poorer survival outcome [24-26]. It was generally accepted that CEA, as an oncofetal antigen, was expressed during fetal life, absent in health adult tissues and re-expressed in cancer tissues[27, 28]. Evidence has shown that forced overexpression of CEA is associated with anoikis, a form of apoptosis caused by detachment from cell matrix, and therefore enhances metastasis[29]. However, Kim et al. demonstrated that CEA mRNA expression in primary tumors is higher than that in liver metastases and inversely correlates with the number of metastatic lymph nodes[30]. Besides, it was reported that it lacked direct proportionality between CEA mRNA and protein expression[31], suggesting that CEA expression was regulated by both transcriptional and post-transcriptional regulation, but the exact mechanisms remained largely unknown. In the present study, we validated CEACAM5 as an oncogene by facilitating tumor migration, and was a direct transcriptional target of HIF1α regulated by FBW7.
Taken together, our study reveals novel role of FBW7 in tumor migration and progression in CRC. Mechanistically, FBW7 regulates CEACAM5 expression in HIF1α-dependent manner, which may offer a potential novel therapeutic avenue in treating CRC.
FBW7 regulates CEACAM5 expression in a HIF1α-dependent manner. The well-known substrates of FBW7, HIF1α and c-Myc, were then transfected into HCT116 and LoVo cells. We found that HIF1α could promote CEACAM5 expression (a), while c-Myc could not (b). (c)Promoter region of CEACAM5 was then analyzed and we found that it harbored one E-box elements AGCGTG from -1 to -5bp. (d) Dual luciferase assay indicated HIF1α and FBW7 altered CEACAM5 promoter activity in a dosage dependent way. When co-transfected with FBW7 and siRNA against HIF1α, FBW7 lost its inhibition effect on CEA promoter. (e) Chromatin immunoprecipitation (ChIP) assay showed that HIF1α occupied the E-boxes in the CEACAM5 promoter region. (f) IHC staining of TMA of colorectal cancer indicated a positive correlation between HIF1α and CEACAM5 (scale bar, 50μm). *P<0.05, **P>0.05.
A proposed model of the mechanism of FBW7-mediated regulation of migration via the HIF1α/CEACAM5 axis in colorectal cancer.
Acknowledgements
This research was supported by the National Science Foundation of China (No. 81702353) and Shanghai Municipal Natural Science Foundation (17ZR1406400). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017
2. Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J. et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology. 2003;124:544-60
3. Li Y, Jia H, Yu W, Xu Y, Li X, Li Q. et al. Nomograms for predicting prognostic value of inflammatory biomarkers in colorectal cancer patients after radical resection. International journal of cancer. 2016;139:220-31
4. Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:2000-15
5. Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature reviews Cancer. 2008;8:83-93
6. Minella AC, Clurman BE. Mechanisms of tumor suppression by the SCF(Fbw7). Cell cycle. 2005;4:1356-9
7. Ji S, Qin Y, Liang C, Huang R, Shi S, Liu J. et al. FBW7 (F-box and WD Repeat Domain-Containing 7) Negatively Regulates Glucose Metabolism by Targeting the c-Myc/TXNIP (Thioredoxin-Binding Protein) Axis in Pancreatic Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:3950-60
8. Cheng Y, Chen G, Martinka M, Ho V, Li G. Prognostic significance of Fbw7 in human melanoma and its role in cell migration. The Journal of investigative dermatology. 2013;133:1794-802
9. Takada M, Zhuang M, Inuzuka H, Zhang J, Zurlo G, Zhang J. et al. EglN2 contributes to triple negative breast tumorigenesis by functioning as a substrate for the FBW7 tumor suppressor. Oncotarget. 2017;8:6787-95
10. Zhang PF, Sheng LL, Wang G, Tian M, Zhu LY, Zhang R. et al. miR-363 promotes proliferation and chemo-resistance of human gastric cancer via targeting of FBW7 ubiquitin ligase expression. Oncotarget. 2016;7:35284-92
11. Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH. et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS letters. 2012;586:1409-18
12. Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nature reviews Cancer. 2014;14:233-47
13. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-7
14. Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J. et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Molecular and cellular biology. 2012;32:2160-7
15. Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36:787-96
16. Zhan P, Wang Y, Zhao S, Liu C, Wang Y, Wen M. et al. FBXW7 negatively regulates ENO1 expression and function in colorectal cancer. Laboratory investigation; a journal of technical methods and pathology. 2015;95:995-1004
17. Li Y, Liang L, Dai W, Cai G, Xu Y, Li X. et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Molecular cancer. 2016;15:55
18. Sinicrope FA, Ruan SB, Cleary KR, Stephens LC, Lee JJ, Levin B. bcl-2 and p53 oncoprotein expression during colorectal tumorigenesis. Cancer research. 1995;55:237-41
19. Zhu JL, Song YX, Wang ZN, Gao P, Wang MX, Dong YL. et al. The clinical significance of mesenchyme forkhead 1 (FoxC2) in gastric carcinoma. Histopathology. 2013;62:1038-48
20. Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T. et al. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer research. 2009;69:3788-94
21. Hagedorn M, Delugin M, Abraldes I, Allain N, Belaud-Rotureau MA, Turmo M. et al. FBXW7/hCDC4 controls glioma cell proliferation in vitro and is a prognostic marker for survival in glioblastoma patients. Cell division. 2007;2:9
22. Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T. et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. International journal of cancer. 2010;126:1828-37
23. Ji S, Qin Y, Shi S, Liu X, Hu H, Zhou H. et al. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell research. 2015;25:561-73
24. Thirunavukarasu P, Sukumar S, Sathaiah M, Mahan M, Pragatheeshwar KD, Pingpank JF. et al. C-stage in colon cancer: implications of carcinoembryonic antigen biomarker in staging, prognosis, and management. Journal of the National Cancer Institute. 2011;103:689-97
25. Thirunavukarasu P, Talati C, Munjal S, Attwood K, Edge SB, Francescutti V. Effect of Incorporation of Pretreatment Serum Carcinoembryonic Antigen Levels Into AJCC Staging for Colon Cancer on 5-Year Survival. JAMA surgery. 2015;150:747-55
26. Becerra AZ, Probst CP, Tejani MA, Aquina CT, Gonzalez MG, Hensley BJ. et al. Evaluating the Prognostic Role of Elevated Preoperative Carcinoembryonic Antigen Levels in Colon Cancer Patients: Results from the National Cancer Database. Annals of surgical oncology. 2016;23:1554-61
27. Krupey J, Gold P, Freedman SO. Physicochemical studies of the carcinoembryonic antigens of the human digestive system. The Journal of experimental medicine. 1968;128:387-98
28. Yan C, Hu Y, Zhang B, Mu L, Huang K, Zhao H. et al. The CEA-/lo colorectal cancer cell population harbors cancer stem cells and metastatic cells. Oncotarget. 2016;7:80700-15
29. Ordonez C, Screaton RA, Ilantzis C, Stanners CP. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer research. 2000;60:3419-24
30. Kim JC, Roh SA, Kim HC, Koo KH, Cho YK, Yu CS. et al. Coexpression of carcinoembryonic antigen and E-cadherin in colorectal adenocarcinoma with liver metastasis. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2003;7:931-8
31. Hauck W, Stanners CP. Control of carcinoembryonic antigen gene family expression in a differentiating colon carcinoma cell line, Caco-2. Cancer research. 1991;51:3526-33
Author contact
![]() Corresponding authors: Sanjun Cai, MD, Ph D. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University.270 Dong'an Road, Xuhui District,Shanghai, 20032 China; Tel.86-021-64175590; E-mail:caisanjun_shcom or Xinxiang Li, MD, Ph D. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University.270 Dong'an Road, Xuhui District, Shanghai, 20032 China; Tel.86-021-64175590; E-mail: lxx1149com
Corresponding authors: Sanjun Cai, MD, Ph D. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University.270 Dong'an Road, Xuhui District,Shanghai, 20032 China; Tel.86-021-64175590; E-mail:caisanjun_shcom or Xinxiang Li, MD, Ph D. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University.270 Dong'an Road, Xuhui District, Shanghai, 20032 China; Tel.86-021-64175590; E-mail: lxx1149com

 Global reach, higher impact
Global reach, higher impact