10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):849-857. doi:10.7150/ijbs.24539 This issue Cite
Research Paper
PRWHMDA: Human Microbe-Disease Association Prediction by Random Walk on the Heterogeneous Network with PSO
1. School of Control Science and Engineering, Shandong University, Jinan, 250061, China.
2. General Clinic, The No. 2 People's Hospital of Tianqiao, Jinan, 250032, China.
3. School of Mathematics and Statistics, Shandong University, Weihai, 264209, China.
Abstract
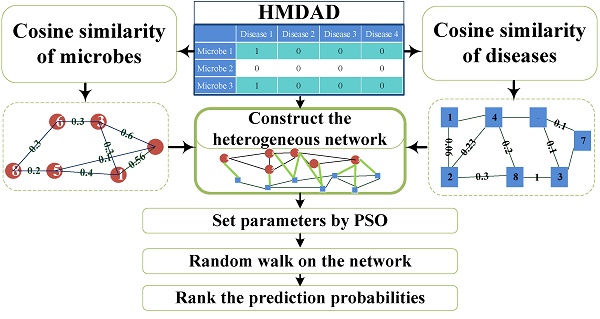
Microorganisms resided in human body play a vital role in metabolism, immune defense, nutrition absorption, cancer control and protection against pathogen colonization. The changes of microbial communities can cause human diseases. Based on the known microbe-disease association, we presented a novel computational model employing Random Walking with Restart optimized by Particle Swarm Optimization (PSO) on the heterogeneous interlinked network of Human Microbe-Disease Associations (PRWHMDA) (see Figure 1). Based on the known human microbe-disease associations, we constructed the heterogeneous interlinked network with Cosine similarity. The extended random walk with restart (RWR) method was derived to get the potential microbe-disease associations. PSO was utilized to get the optimal parameters of RWR. To evaluate the prediction effectiveness, we performed leave one out cross validation (LOOCV) and 5-fold cross validation (CV), which got the AUC (The area under ROC curve) of 0.915 (LOOCV) and the average AUCs of 0.8875 ± 0.0046 (5-fold CV). Moreover, we carried out three case studies of asthma, inflammatory bowel disease (IBD) and type 1 diabetes (T1D) for the further evaluation. The result showed that 10, 10 and 9 of top-10 predicted microbes were verified by previously published experimental results, respectively. It is anticipated that PRWHMDA can be effective to identify the disease-related microbes and maybe helpful to disclose the relationship between microorganisms and their human host.
Keywords: disease-microbe association prediction, random walk with restart, particle swarm optimization.

 Global reach, higher impact
Global reach, higher impact