10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):858-862. doi:10.7150/ijbs.24581 This issue Cite
Research Paper
CRISPRMatch: An Automatic Calculation and Visualization Tool for High-throughput CRISPR Genome-editing Data Analysis
1. Jiangsu Key Laboratory of Crop Genetics and Physiology, Co-Innovation Centre for Modern Production Technology of Grain Crops, Key Laboratory of Plant Functional Genomics of the Ministry of Education, Yangzhou University, Yangzhou 225009, China
2. Department of Biotechnology, School of Life Science and Technology, Centre for Informational Biology, University of Electronic Science and Technology of China, Chengdu 610054, China.
3. Joint International Research Laboratory of Agriculture and Agri-Product Safety, the Ministry of Education of China, Yangzhou University, Yangzhou 225009, China
Abstract
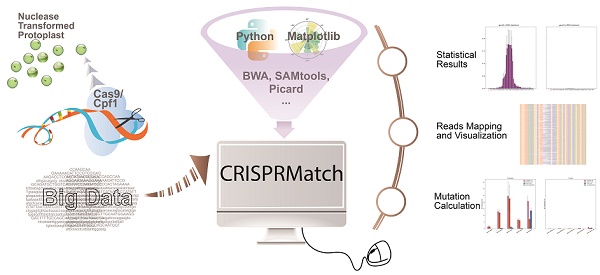
Custom-designed nucleases, including CRISPR-Cas9 and CRISPR-Cpf1, are widely used to realize the precise genome editing. The high-coverage, low-cost and quantifiability make high-throughput sequencing (NGS) to be an effective method to assess the efficiency of custom-designed nucleases. However, contrast to standardized transcriptome protocol, the NGS data lacks a user-friendly pipeline connecting different tools that can automatically calculate mutation, evaluate editing efficiency and realize in a more comprehensive dataset that can be visualized. Here, we have developed an automatic stand-alone toolkit based on python script, namely CRISPRMatch, to process the high-throughput genome-editing data of CRISPR nuclease transformed protoplasts by integrating analysis steps like mapping reads and normalizing reads count, calculating mutation frequency (deletion and insertion), evaluating efficiency and accuracy of genome-editing, and visualizing the results (tables and figures). Both of CRISPR-Cas9 and CRISPR-Cpf1 nucleases are supported by CRISPRMatch toolkit and the integrated code has been released on GitHub (
Keywords: CRISPR, NGS data, automatic pipeline, mutation calculation, genome-editing efficiency

 Global reach, higher impact
Global reach, higher impact