10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(9):1787-1801. doi:10.7150/ijbs.32142 This issue Cite
Research Paper
SWPepNovo: An Efficient De Novo Peptide Sequencing Tool for Large-scale MS/MS Spectra Analysis
1. College of Information Science and Engineering, Hunan University, Changsha, China;
2. College of Information Science and Engineering, Hunan University, National Supercomputing Center in Changsha, Changsha, China.
3. College of Information Science and Engineering, Hunan University, Department of Computer Science, State University of New York, NY, USA.
4. State Key Laboratory of Mathematic Engineering and Advance Computing, Wuxi Jiangnan Institute of Computing Technology, Jiangsu, China.
5. School of Computer Science and Engineering, Nanyang Technological University, Singapore.
Abstract
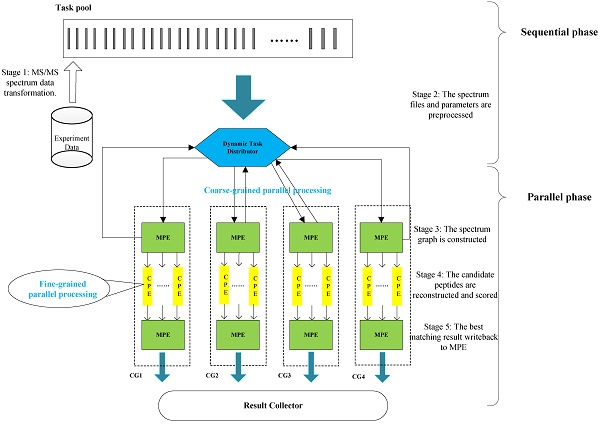
Tandem mass spectrometry (MS/MS)-based de novo peptide sequencing is a powerful method for high-throughput protein analysis. However, the explosively increasing size of MS/MS spectra dataset inevitably and exponentially raises the computational demand of existing de novo peptide sequencing methods, which is an issue urgently to be solved in computational biology. This paper introduces an efficient tool based on SW26010 many-core processor, namely SWPepNovo, to process the large-scale peptide MS/MS spectra using a parallel peptide spectrum matches (PSMs) algorithm. Our design employs a two-level parallelization mechanism: (1) the task-level parallelism between MPEs using MPI based on a data transformation method and a dynamic feedback task scheduling algorithm, (2) the thread-level parallelism across CPEs using asynchronous task transfer and multithreading. Moreover, three optimization strategies, including vectorization, double buffering and memory access optimizations, have been employed to overcome both the compute-bound and the memory-bound bottlenecks in the parallel PSMs algorithm. The results of experiments conducted on multiple spectra datasets demonstrate the performance of SWPepNovo against three state-of-the-art tools for peptide sequencing, including PepNovo+, PEAKS and DeepNovo-DIA. The SWPepNovo also shows high scalability in experiments on extremely large datasets sized up to 11.22 GB. The software and the parameter settings are available at
Keywords: Large-scale MS/MS spectra analysis, de novo peptide sequencing, high performance computing, SW26010

 Global reach, higher impact
Global reach, higher impact