10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(3):495-503. doi:10.7150/ijbs.39582 This issue Cite
Research Paper
NKILA, a prognostic indicator, inhibits tumor metastasis by suppressing NF-κB/Slug mediated epithelial-mesenchymal transition in hepatocellular carcinoma
1. Department of Surgery, Division of Hepatobiliary and Pancreatic Surgery, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310000, China.
2. NHFPC Key Laboratory of Combined Multi-organ Transplantation, Hangzhou 310000, China.
3. Key Laboratory of the diagnosis and treatment of organ Transplantation, CAMS.
4. Key Laboratory of Organ Transplantation, Zhejiang Province, Hangzhou 310003, China.
5. Collaborative innovation center for Diagnosis treatment of infectious diseases, Hangzhou 310000, China.
*These authors contributed equally to this work.
Received 2019-8-23; Accepted 2019-11-24; Published 2020-1-1
Abstract
The metastasis of hepatocellular carcinoma (HCC) is one of the major obstacles hindering its therapeutic efficacy, leading to low surgical resection rate, high mortality and poor prognosis. Accumulating evidence has shown that both long noncoding RNA (lncRNA) and NF-κB play vital roles in the regulation of cancer metastasis. However, the clinical significance and biological function of NKILA (NF-κB interacting lncRNA) and its interaction with NF-κB in HCC remain unknown. In this study, we demonstrated that NKILA was down-regulated in HCC tissues and cell lines, and decreased NKILA expression was significantly associated with larger tumor size and positive vascular invasion in HCC patients. NKILA reduction was an independent risk factor of HCC patients' poor prognosis, and the 5-year overall survival (OS) rates of patients with low and high NKILA expression were 15.6% and 60.0%, respectively. Moreover, NKILA inhibits migration and invasion of HCC cells both in vitro and in vivo. Mechanistically, NKILA prevents Slug/epithelial to mesenchymal transition (EMT) pathway via suppressing phosphorylation of IκBα, p65 nuclear translocation and NF-κB activation. In conclusion, these results indicate that NKILA might serve as an effective prognostic biomarker and a promising therapeutic target against HCC metastasis.
Keywords: LncRNA-NKILA, NF-κB, Hepatocellular carcinoma, Metastasis, Epithelial to mesenchymal transition
Introduction
Hepatocellular carcinoma (HCC) is an urgent public health problem. Worldwide, HCC is the third leading cause of cancer-related mortality [1]. In China, HCC is one of the most common malignancies and the top cause of cancer-related death in men under 60 [2]. The metastasis of HCC is one of the major obstacles hindering its therapeutic efficacy, leading to low surgical resection rate, high mortality and poor prognosis. The median survival and one-year survival rate of patients with extra-hepatic metastasis from HCC are only 4.9-7 months and 21.7%-24.9% [3, 4]. Therefore, exploring the molecular mechanism of HCC metastasis is urgently needed to develop new diagnostic and therapeutic strategies to improve the situation.
Long noncoding RNAs (lncRNAs) refer to a class of transcripts longer than 200 nucleotides without protein-coding ability [5]. Currently, many studies have revealed that lncRNAs are frequently dysregulated in multiple cancers and involved in various biological functions, such as proliferation, metastasis, apoptosis and drug resistance [6-8]. Particularly, extensive evidence has shown that lncRNAs have an important part in regulating HCC metastasis. For instance, lncRNA miR503HG inhibits HCC metastasis by regulating the HNRNPA2B1/NF-κB pathway [9]. lncRNA ZFAS1 promotes metastasis via sponging miR-150 and up-regulating ZEB1 in hepatocellular carcinoma [10]. Researches focusing on lncRNA provide a new perspective for further elucidation of the molecular mechanism underlying HCC metastasis.
Nuclear factor-κB (NF-κB) is a family of transcription factors critical in regulating immune and inflammatory responses, cancer initiation and progression [11, 12]. Normally, NF-κB p65/p50 components associated with IκB are retained in the cytoplasm as an inactive state. Upon stimulation, IκB kinase complex (IKK) is activated and phosphorylates IκBα, leading to its ubiquitination and subsequent degradation. Consequently, p65/p50 heterodimer is released and translocated to the nucleus, regulating the expression of genes it binds to [13, 14]. To date, accumulating studies have shown that NF-κB plays vital roles in the regulation of epithelial-mesenchymal transition (EMT) and cancer metastasis [15, 16]. A better understanding of the specific function of NF-κB pathway on HCC metastasis is essential to accelerate the development of novel anti-metastasis therapies. NF-κB interacting lncRNA (NKILA) has been reported to inhibit NF-κB pathway and suppress tumor metastasis in breast cancer [17] and non-small cell lung cancer [18]. However, the precise role of NKILA and its interaction with NF-κB in HCC remain unclear.
Therefore, we detected the expression pattern of NKILA in HCC tissues and investigated the function and mechanism of NKILA on HCC cell biological behavior. Herein, we demonstrate that NKILA is down-regulated in HCC tissues and cell lines, and positively correlates with HCC patients' overall survival. Moreover, NKILA inhibits metastasis of HCC cells both in vitro and in vivo. Mechanistically, NKILA prevents EMT via the inhibition of NF-κB/Slug pathway. These results indicate that NKILA might serve as an effective prognostic biomarker and a promising therapeutic target against HCC metastasis.
Methods
Patient specimens
139 pairs of tumor and corresponding adjacent normal tissues were collected from HCC patients who underwent curative hepatectomy at the First Affiliated Hospital, Zhejiang University School of Medicine from 2011 to 2013. All included individuals received no chemotherapy or radiation therapy before surgery. The end point of follow up was Dec. 2018. Written informed consent was obtained from all subjects, and this study was approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine.
Cell culture
All the cells were purchased from China Center for Type Culture Collection (CCTCC). Human hepatocellular carcinoma cells (SMMC-7721, SK-hep-1, HCC-LM3, Huh-7) and human immortalized normal hepatocytes (L-02) [19] were maintained in DMEM (Sigma, USA), MEM (Gibco, USA), or RPMI 1640 (Sigma, USA) containing 10% fetal calf serum (Gibco, USA) at 37 °C in a humidified incubator (Thermo Scientific, USA) with 5% CO2.
RNA isolation and qRT-PCR
Total RNA was extracted from tissue samples or cell lines using TRIzol reagent (Invitrogen, USA) following the manufacturer's protocol. Then the reverse transcription was performed using the iScript™ cDNA Synthesis Kit (Bio-Rad, China) according to the manufacturer's instruction. The expression level of NKILA was measured by SYBR green qRT-PCR assay (Takara, China). The primers (Tsingke, China) were as follows: NKILA forward 5'-AACCAAACCTACCACAACG-3' and reverse 5'-ACCACTAAGTCAATCCCAGGTG-3'; GAPDH forward 5'- CCTGGTATGACAACGAATTTG-3' and reverse 5'-CAGTGAGGGTCTCTCTCTTCC-3'.
Lentivirus transfection
SMMC-7721 and HCC-LM3 were transfected with lentiviral vector expressing NKILA or empty vector (Genechem, China) following standard procedure. 48 h after transfection, the cells were treated with puromycin (Sigma, USA) at a final concentration of 4 μg/ml for 2 weeks to establish stable expression cell lines.
Cell proliferation assays
Cell Counting Kit-8 (Dojindo Laboratories, Japan) was used to detect cell proliferation. Cells (1×104) were seeded into 96-well plates and after one night of incubation, determined at 450 nm using a microplate reader (Bio-Rad, CA) for four consecutive days.
Transwell migration and invasion assays
Migration assays were performed using 24-well Transwell chambers (Corning, USA). 5×104 cells resuspended in 200 μL serum-free medium were seeded to the upper chamber. The lower chamber was filled with 500 μL medium containing 10% FBS. After incubation for 24 h, cells on the membrane were stained with crystal violet (Thermo Scientific, USA) and then were examined and photographed using an inverted microscope (Leica, USA). Invasion assays were performed according to the same procedures except that transwell chamber was coated with 50 μL matrigel and incubation for 48 h.
In vivo metastasis assay
A total of 106 cells in 100 μL PBS were injected into each athymic nude mice through tail veins to establish in vivo metastasis models. After 6 weeks, the animals were sacrificed and the lungs were harvested and fixed in formalin. After embedded with paraffin, slides were prepared and underwent hematoxylin and eosin (H&E) staining. Afterwards, the stained slides were examined and photographed under microscopy. The animal experiments were approved by the Ethics Committee for Laboratory Animals of the First Affiliated Hospital, Zhejiang University School of Medicine.
Western blot analysis and antibodies and subcellular extraction
The detailed procedure has been described in our previous study [20]. Briefly, proteins were isolated with RIPA lysis buffer (Servicebio, China) and quantified with BCA Protein assay kit (Thermo Scientific, USA). Then equal amounts of proteins were fractionated on 10% SDS-PAGE gels (Invitrogen, USA) and transferred to PVDF membranes (Millipore, USA). After blocked with skim milk, the membranes were incubated with various primary antibodies at 4 °C overnight, and then incubated with corresponding secondary antibodies for 1h. Subsequently, the bands were visualized using ECL kits (Abcam, USA). The primary antibodies (Cell Signaling Technology, USA) were as follows: E-Cadherin (#3195), N-Cadherin (#13116), Vimentin (#5741), Slug (#9585), β-actin (#4970), p-IKKα/β (#2697), p-IκBα (#2859), IκBα (#4814), p65 (#8242), p-p65 (#3033), Lamin-A (#86846).
Subcellular fractions were performed using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, China) following the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using SPSS version 22.0 (SPSS, USA). Student-t test or one-way ANOVA was used to compare the difference between groups. All the experiments were performed at least 3 times and each value was presented as mean±S.D. The relationship between NKILA expression and clinicopathological characteristics were analyzed by Chi-squared test, and survival analysis was performed using Kaplan-Meier curves and log-rank test. Cox proportional hazards model was used to analyze OS predictors. Difference was considered significant at a level of P < 0.05.
Results
NKILA is down-regulated in HCC and acts as an independent predictor of HCC patients' prognosis
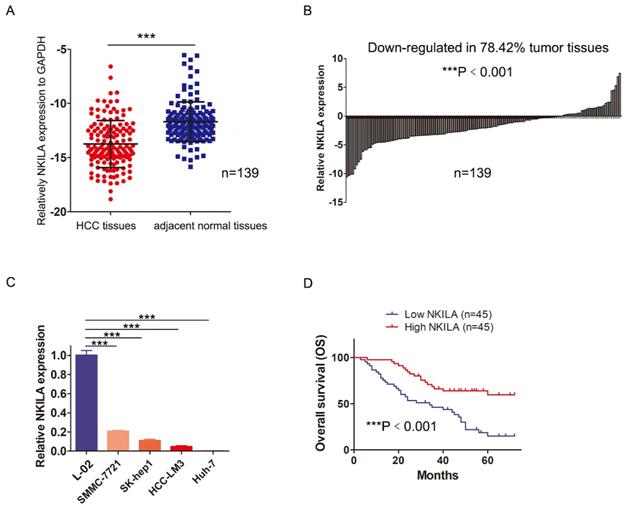
In order to assess the role of NKILA in HCC, we first measured the expression of NKILA in 139 pairs of HCC and corresponding adjacent normal tissues by qRT-PCR. As shown in Figure 1A, the expression level of NKILA significantly decreased in HCC tissues (P < 0.001). Compared with corresponding adjacent normal tissues, down-regulation of NKILA expression was observed in 78.42% (109/139) of HCC tissues (P < 0.001, Figure 1B). Moreover, the expression level of NKILA was remarkably lower in four human HCC cell lines than human immortalized normal hepatocytes L-02 (P < 0.001, Figure 1C).
To explore the clinicopathological significance of NKILA, 90 out of 139 patients were taken into analysis (49 patients with incomplete clinicopathological data or lost to follow-up within 2 years after surgery were excluded). As depicted in Table 1, chi-square analysis revealed that decreased NKILA expression in HCC was significantly associated with larger tumor size and positive vascular invasion. Kaplan-Meier curves and log-rank test showed that the overall survival (OS) of the patients with low NKILA expression was significantly shorter than those with high NKILA expression (P < 0.001, Figure 1D). The 5-year OS rates of patients with low and high NKILA expression were 15.6% and 60.0%, respectively. Moreover, as shown in Table 2, univariate and multivariate analysis indicated that NKILA expression (HR 0.325, 95% CI 0.181-0.582, P = 0.009) as well as AFP values and tumor differentiation grade, were independent predictors of OS in HCC patients.
In a word, NKILA is down-regulated in HCC tissues and cell lines, and serves as an independent predictor of HCC patients' overall survival.
NKILA inhibits migration and invasion of HCC cells in vitro
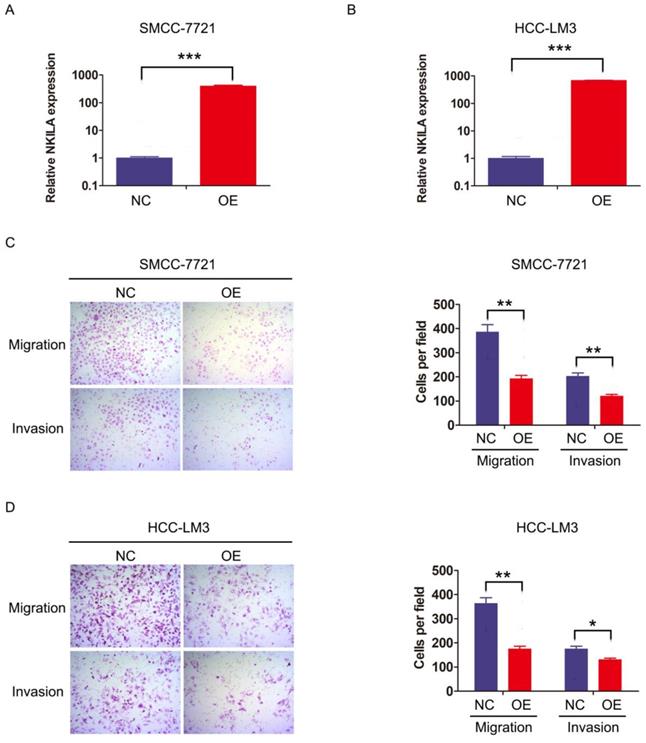
We then investigated the function of NKILA on HCC cell biological behavior. SMMC-7721 and HCC-LM3 were transfected with lentiviral vector expressing NKILA or empty vector as negative control to establish stable expression cell lines (Figure 2A-2B). CCK-8 assay showed that overexpression of NKILA had little influence on the proliferation of these two HCC cell lines compared with negative control (Figure S1). Notably, as depicted in transwell assay, overexpression of NKILA significantly suppressed the migration and invasion abilities of SMMC-7721 and HCC-LM3 in vitro (Figure 2C-2D).
NKILA is down-regulated in HCC and acts as an independent predictor of HCC patients' prognosis. (A) The expression of NKILA in 139 pairs of HCC tissues and corresponding adjacent normal tissues was detected by qRT-PCR. (B) The expression of NKILA in HCC tissues was normalized to that of corresponding noncancerous tissues. The data was shown as log2(Fold change) = log2(TNKILA/NNKILA). (C) NKILA expression in human immortalized normal hepatocytes L-02 and four human HCC cell lines was detected by qRT-PCR. (D) Kaplan-Meier overall survival curves of 90 HCC patients with low and high NKILA levels. The data was presented as mean ± SD of three independent experiments. ***P < 0.001.
Correlation analysis between NKILA expression and clinicopathological parameters of HCC patients
| Variables | NKILA expression | P value | |
|---|---|---|---|
| low | high | ||
| Age (years) | 0.829 | ||
| ≤55 | 17 | 18 | |
| >55 | 28 | 27 | |
| Gender | 0.292 | ||
| Female | 6 | 3 | |
| Male | 39 | 42 | |
| HBV | 0.803 | ||
| Absent | 10 | 11 | |
| Present | 35 | 34 | |
| AFP (ng/ml) | 0.090 | ||
| ≤400 | 16 | 24 | |
| >400 | 29 | 21 | |
| Cirrhosis | 0.455 | ||
| Absent | 12 | 9 | |
| Present | 33 | 36 | |
| Tumor Size (cm) | 0.006** | ||
| ≤5 | 19 | 32 | |
| >5 | 26 | 13 | |
| Tumor number | 0.822 | ||
| =1 | 31 | 30 | |
| >1 | 14 | 15 | |
| Vascular invasion | 0.003** | ||
| Negative | 25 | 38 | |
| Positive | 20 | 7 | |
| Tumor differentiation | 0.393 | ||
| Poor | 28 | 24 | |
| Moderate-Well | 17 | 21 | |
HCC: hepatocellular carcinoma; HBV: hepatitis B virus; AFP: alpha fetoprotein. **P < 0.01.
Univariate and multivariate analysis of clinicopathological parameters related to overall survival in HCC patients
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (≤55/ >55) | 1.268 (0.721-2.231) | 0.409 | - | - |
| Gender (Female/Male) | 0.858 (0.366-2.011) | 0.725 | - | - |
| HBV (Absent/Present) | 0.901 (0.480-1.692) | 0.746 | - | - |
| AFP (>400/≤400) | 3.949 (2.062-7.560) | 0.000*** | 3.000 (1.549-5.810) | 0.001** |
| Cirrhosis (Absent/Present) | 0.918 (0.481-1.750) | 0.795 | - | - |
| Tumor Size (>5cm/≤5cm) | 1.861 (1.077-3.215) | 0.026* | - | - |
| Tumor number (=1/>1) | 0.599 (0.344-1.043) | 0.070 | - | - |
| Vascular invasion (Positive/Negative) | 2.779 (1.577-4.900) | 0.000 | - | - |
| Tumor differentiation (Moderate-Well/Poor) | 0.375 (0.204-0.688) | 0.002** | 0.463 (0.252-0.852) | 0.013* |
| NKILA expression (High/Low) | 0.325 (0.181-0.582) | 0.000*** | 0.454 (0.251-0.822) | 0.009** |
HCC: hepatocellular carcinoma; HBV: hepatitis B virus; AFP: alpha fetoprotein.
*P < 0.05, **P < 0.01, ***P < 0.001.
NKILA suppresses metastasis of HCC in vivo
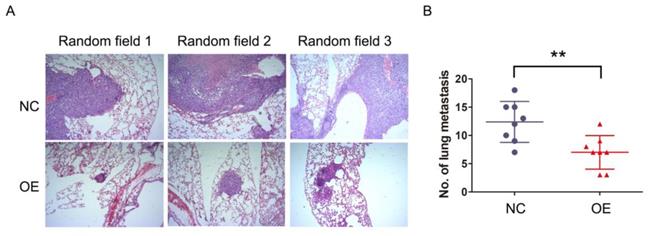
The previous results demonstrated that NKILA inhibited migration and invasion of HCC cells in vitro, which raised a concern regarding whether this phenomenon would occur in vivo as well. SMMC-7721 cells stably expressing NKILA or empty vector as established before were injected into the tail vein of athymic nude mice to produce in vivo metastasis models. After 6 weeks, the lungs were examined by H&E staining. As shown in Figure 3A-3B, the number and size of metastatic nodules showed remarkable reduction in the overexpression group compared with negative control group. Taken together, overexpression of NKILA could also significantly suppress metastasis of HCC in vivo.
NKILA inhibits migration and invasion of HCC cells in vitro. (A-B) The expression of NKILA in SMMC-7721 and HCC-LM3 cells transfected with lentiviral vector expressing NKILA (overexpression group) or empty vector (negative control group) was detected by qRT-PCR. (C-D) The migration and invasion abilities of SMMC-7721 and HCC-LM3 cells in NKILA overexpression and negative control groups were detected by transwell assay, and images were obtained at 400× magnification. The data was presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. OE, overexpression; NC, negative control.
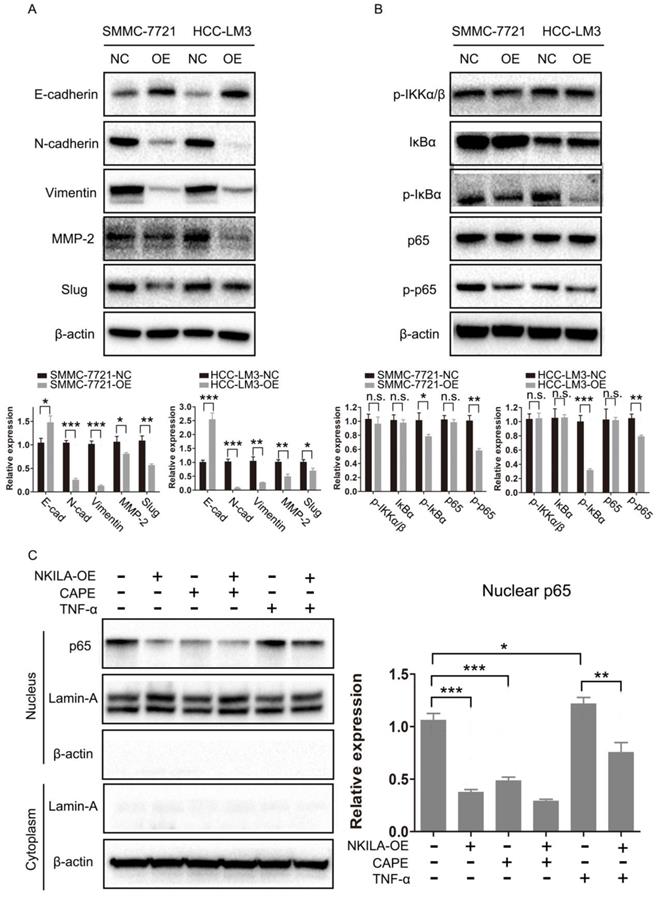
NKILA prevents EMT via the inhibition of NF-κB/Slug pathway in HCC
As EMT is closely related with metastasis of cancer cells [21], we further studied the connection of NKILA and EMT in HCC. As shown in Figure 4A, overexpression of NKILA led to a notable increase in the expression of E-Cadherin and a decrease in the expression of N-Cadherin, Vimentin, MMP-2 and Slug, indicating that NKILA could prevent EMT. NF-κB pathway was reported to play a pivotal part in regulating EMT of cancer cells [15, 16]. Therefore, we next explored the role of NF-κB in NKILA induced EMT suppression. As depicted in Figure 4B, overexpression of NKILA had little effect on the expression of p-IKKα/β, but significantly decreased the expression of p-IκBα and p-p65. In addition, we tested p65 nuclear translocation to reflect the NF-κB activity. A classical NF-κB inducer TNF-α [22] and an inhibitor CAPE [23] were used to treat SMMC-7721 cells. As shown in Figure 4C, overexpression of NKILA or treated with CAPE remarkably inhibited p65 nuclear translocation compared with negative control, while treated with TNF-α promoted the translocation. Moreover, overexpression of NKILA could substantially withdraw the TNF-α induced nuclear translocation of p65. Collectively, NKILA prevents EMT via the inhibition of NF-κB/Slug pathway in HCC (Figure 5).
NKILA suppresses metastasis of HCC in vivo. (A) Representative images of lung sections stained by H&E in each group of SMMC-7721 cells. (B) The numbers of metastatic lesions in the lungs at 6 weeks after tail vein injection. **P < 0.01. OE, NKILA overexpression; NC, negative control.
Discussion
The activating invasion and metastasis is one of the six hallmarks of cancer [24]. In HCC, intrahepatic and extrahepatic metastases are the major reasons leading to high recurrence rate and poor prognosis [25]. This highlights the urgency of identifying high-risk patients in advance and establishing promising therapeutic targets for successful intervention. Nowadays, emerging studies have shown that lncRNAs are crucial for HCC metastasis and prognostic prediction. lncRNA GAS5 functions as a tumor suppressor in HCC metastasis through directly interacting with miR-182 and low level of GAS5 correlates with poor outcomes [26]. lncRNA GALH promotes HCC metastasis via epigenetically regulating Gankyrin and acts as an independent unfavorable prognostic indicator [27]. In this study, we focused on the newly identified lncRNA NKILA, evaluated the clinicopathological significance of NKILA in HCC patients and investigated the function and mechanism of NKILA on HCC cell biological behavior.
NKILA was first identified as an lncRNA upregulated by inflammatory cytokines via NF-κB Signaling in breast cancer. It interacts with NF-κB/IκB to form a stable complex, and directly masks phosphorylation motifs of IκB, thereby inhibiting IKK-induced IκB phosphorylation and NF-κB activation. Furthermore, NKILA suppresses breast cancer invasiveness, and NKILA expression was an independent predictor of breast cancer patients' prognosis [17]. As NF-κB is a critical link between liver inflammation and cancer [28, 29], it is essential to figure out the precise role of NKILA and its interaction with NF-κB in HCC. To the best of our knowledge, this is the first study demonstrating the clinical significance of NKILA in HCC patients and investigating the function of NKILA on HCC cell biological behavior. Our results showed that the expression of NKILA was down-regulated in HCC tissues and decreased NKILA expression was significantly associated with larger tumor size and positive vascular invasion. NKILA reduction was an independent risk factor of HCC patients' poor prognosis. Moreover, overexpression of NKILA suppressed the migration and invasion of HCC cells both in vitro and in vivo by inhibiting the phosphorylation of IκB and NF-κB pathway. However, as the expression of NKILA was relatively low in HCC cell lines, we failed to do the knock-down experiments using siRNA and CRISPR/Cas9, therefore we only included the gain-of-function assays in this study.
Epithelial to mesenchymal transition (EMT) refers to a multistep, plastic and reversible morphologic transformation that enables epithelial cancer cells to acquire mesenchymal features, which has a significant role in tumor progression and metastasis [30]. A set of transcriptional factors, including Snail, Slug, Twist, Zeb1 and Zeb2, orchestrate the EMT and subsequent migratory process [31]. In this study, we demonstrated that the molecular mechanism underlying the effect of NKILA on HCC metastasis was through inhibiting Slug-EMT pathway. Overexpression of NKILA resulted in a notable increase in the expression of E-Cadherin and a decrease in the expression of N-Cadherin, Vimentin, MMP-2 and Slug. Our data contributed to a better understanding of the complex regulatory network of EMT.
In summary, NKILA is down-regulated in HCC tissues and cell lines, and positively correlates with HCC patients' overall survival. Moreover, NKILA inhibits HCC metastasis both in vitro and in vivo by hindering IκB phosphorylation and NF-κB activation, and subsequently suppressing Slug regulated EMT. These results indicate that NKILA might serve as an effective prognostic biomarker and a promising therapeutic target against HCC metastasis.
NKILA prevents EMT via the inhibition of NF-κB/Slug pathway in HCC. (A-B) EMT and NF-κB pathway related proteins of SMMC-7721 and HCC-LM3 cells in NKILA overexpression and negative control groups were detected by western blot. Relative semi‑quantitative analysis results were presented below. (C) Nuclear p65 of SMMC-7721 cells in NKILA overexpression and negative control groups treated with or without TNF-α or CAPE was detected by western blot. β-actin and Lamin-A were the loading control for cytoplasm and nuclear, respectively. Relative semi‑quantitative analysis results were presented (right panel). n.s., P > 0.05; *P < 0.05, **P < 0.01, ***P < 0.001. OE, overexpression; NC, negative control.
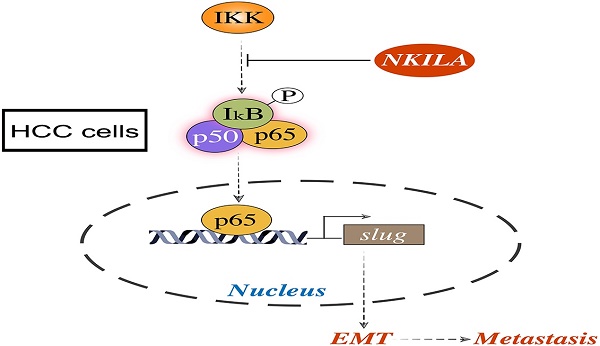
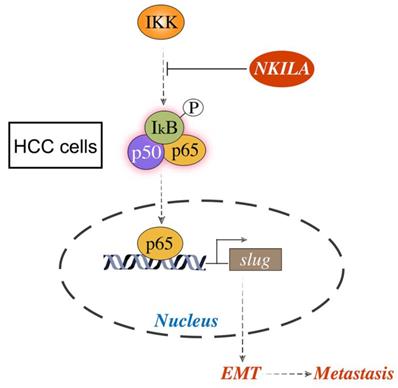
Schematic model of the underlying molecular mechanism of NKILA on HCC metastasis. NKILA inhibits HCC cells migration and invasion by hindering IKK induced IκB phosphorylation, p65 nuclear translocation and NF-κB activation, and subsequently suppressing Slug regulated EMT.
Abbreviations
AFP: alpha fetoprotein; CAPE: caffeic acid phenethyl ester; EMT: epithelial to mesenchymal transition; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; H&E: hematoxylin and eosin; LncRNA: long noncoding RNA; NC: negative control; NF-κB: Nuclear factor-κB; NKILA: NF-κB interacting lncRNA; OE: overexpression; OS: overall survival.
Supplementary Material
Supplementary figure S1.
Acknowledgements
This research was supported by Innovative Research Groups of National Natural Science Foundation of China (No. 81721091); National S&T Major Project (No. 2017ZX10203205); Zhejiang International Science and Technology Cooperation Project (NO.2016C04003).
Ethics Committee Approval and Patient Consent
The animal experiments in this study were approved by the Ethics Committee for Laboratory Animals of the First Affiliated Hospital, Zhejiang University School of Medicine. All patient specimens were obtained with written informed consent, and approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66:115-32
3. Aino H, Sumie S, Niizeki T, Kuromatsu R, Tajiri N, Nakano M. et al. Clinical characteristics and prognostic factors for advanced hepatocellular carcinoma with extrahepatic metastasis. Molecular and clinical oncology. 2014;2:393-8
4. Natsuizaka M, Omura T, Akaike T, Kuwata Y, Yamazaki K, Sato T. et al. Clinical features of hepatocellular carcinoma with extrahepatic metastases. Journal of gastroenterology and hepatology. 2005;20:1781-7
5. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-6
6. Liu Q, Yu W, Zhu S, Cheng K, Xu H, Lv Y. et al. Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR-18a-5p. Journal of cellular physiology. 2018;234:757-68
7. Wang M, Guo C, Wang L, Luo G, Huang C, Li Y. et al. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell death & disease. 2018;9:238
8. Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y. et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell death & disease. 2018:9
9. Wang H, Liang L, Dong Q, Huan L, He J, Li B. et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-kappaB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814-29
10. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z. et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer research. 2015;75:3181-91
11. Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Molecular Cancer. 2013;12:86
12. Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-kappaB signalling circuit. Immunology. 2017;150:379-88
13. Hayden HS, Ghosh SK. Signaling to NF-kappaB. Genes Dev. 2004;18:2195
14. Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685-705
15. Huber MA, Ninel A, Bernd B, Stefan G, Andreas S, Hubert P. et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. Journal of Clinical Investigation. 2004;114:569-81
16. Huber MA, Beug H, Wirth T. Epithelial-Mesenchymal Transition: NF-κB Takes Center Stage. Cell Cycle. 2004;3:1477-80
17. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370-81
18. Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J. et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. Journal of experimental & clinical cancer research: CR. 2017;36:54
19. Hu X, Yang T, Li C, Zhang L, Li M, Huang W. et al. Human Fetal Hepatocyte Line, L-02, Exhibits Good Liver Function In Vitro and in an Acute Liver Failure Model. Transplantation Proceedings. 45: 695-700.
20. Chen R, Cheng Q, Owusu-Ansah KG, Chen J, Song G, Xie H, Zhou L, Xu X, Jiang D, Zheng S. Cabazitaxel, a novel chemotherapeutic alternative for drug-resistant hepatocellular carcinoma. Am J Cancer Res. 2018;8(7):1297-1306
21. Yang J, Weinberg RA. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Developmental Cell. 2008;14:818-29
22. Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-kappaB addiction and its role in cancer: 'one size does not fit all'. Oncogene. 2011;30:1615-30
23. Natarajan K, Singh S, Burke TR, Grunberger D, Aggarwal BB. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9090-5
24. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
25. Budhu AS, Forgues M, Ye QH, Jia HL, He P, Zanetti K. et al. Prediction of metastatic potential, recurrence and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer cell. 2006;10:99-111
26. Chen Li Y, Li M Wang L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Am J Cancer Res. 2019;9(1):108-121
27. Xu X, Lou Y, Tang J, Teng Y, Zhang Z, Yin Y. et al. The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell death & disease. 2019;10:86
28. Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T. et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology. 2014;60:1659-73
29. Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang S-M. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Molecular Cancer. 2014;13:35 -
30. Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21-45
31. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nature reviews Cancer. 2018;18:128-34
Author contact
![]() Corresponding authors: Jian Wu, drwujianedu.cn & Shusen Zheng, shusenzhengedu.cn.
Corresponding authors: Jian Wu, drwujianedu.cn & Shusen Zheng, shusenzhengedu.cn.

 Global reach, higher impact
Global reach, higher impact