Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(12):2094-2103. doi:10.7150/ijbs.44420 This issue Cite
Review
Emerging Roles of Long non-coding RNAs in The Tumor Microenvironment
1. Taizhou University hospital, Taizhou University, Taizhou, Zhejiang, 318000, China
2. Taizhou Municipal Hospital, Taizhou University, Taizhou, Zhejiang, 318000, China
*These authors contributed equally to this work
Received 2020-3-10; Accepted 2020-4-13; Published 2020-5-18
Abstract
Long non-coding RNAs (lncRNAs) are a diverse class of longer than 200 nucleotides RNA transcripts that have limited protein coding capacity. LncRNAs display diverse cellular functions and widely participate in both physiological and pathophysiological processes. Aberrant expressions of lncRNAs are correlated with tumor progression, providing sound rationale for their targeting as attractive anti-tumor therapeutic strategies. Emerging evidences support that lncRNAs participate in tumor-stroma crosstalk and stimulate a distinctive and suitable tumor microenvironment (TME). The TME comprises several stromal cells such as cancer stem cells (CSCs), cancer-associated endothelial cells (CAEs), cancer-associated fibroblasts (CAFs) and infiltrated immune cells, all of which are involved in the complicated crosstalk with tumor cells to affect tumor progression. In this review, we summarize the essential properties and functional roles of known lncRNAs in related to the TME to validate lncRNAs as potential biomarkers and promising anti-cancer targets.
Keywords: long non-coding RNA, tumor microenvironment, immune cells, cancer-stem cells, cancer-associated endothelial cells, cancer-associated fibroblasts
Introduction
Long noncoding RNAs (lncRNAs) are a large class of transcribed RNA molecules that are greater than 200 nucleotides in length and have limited protein coding potential for the lack of functional open readings (1, 2). In term of functions of lncRNAs, guide, decoy, signaling and scaffold functions have been identified (3). Guide lncRNAs modulate gene expression via recruiting chromatin modifying enzymes to specific genomic regions, while decoy lncRNAs sequester transcription factors away from chromatin. Signaling lncRNAs serve as molecular signals and integrate with specific signaling pathways or events. Scaffold lncRNAs act as platforms to recruit multiple proteins to assemble functional units, such as ribonucleoprotein complexes, and regulate gene expression. To date, lncRNAs have been found to actively participate in the regulation of various aspects of tumor development, including growth, metastasis and recurrence (4-6). Indeed, lncRNAs can function as oncogenes or tumor suppressors, and aberrant expressions are linked to various human cancers, such as prostate cancer (7), colorectal cancer (8) and hepatocellular carcinoma (9). Notably, like circulating miRNAs, lncRNAs can also be detected in the sputum, blood, and urine of cancer patients, indicating that some lncRNAs may be potential non-invasive diagnosis targets for human cancers. For example, lncRNA PCA3, specifically expressed in the prostate, has been developed as an early diagnosis marker of prostate cancer, which is more sensitive and specific than serum prostate specific antigen (PSA) (10, 11). LncRNAs are also being prime targets for cancer therapy (5). LncRNAs appear to form a secondary structure and act as multicomponent complexes, making lncRNAs attractive for cancer therapeutic intervention (12). In addition, several other features, such as low abundance and tissue specificity, also support lncRNAs as potential efficacious anti-tumor targets. LncRNA H19, upregulated in many cancers, exerts oncogenic effects through promoting cancer progression, angiogenesis, and metastasis (13, 14). H19-DTA (BC-819) is a DNA plasmid that carries the diphtheria toxin gene under the regulation of the H19 promoter sequence and therefore has a potential therapeutic effect on a variety of tumors overexpressing H19 gene. Currently, BC-819 has been advanced into a series of phase I/IIb clinical trials for patients suffering from bladder, ovarian, and pancreatic cancer (https://www.clinicaltrials.gov/), and four completed clinical trials have demonstrated BC-819 is safe and feasible for tumor treatment (15-17). In short, lncRNAs are emerging as promising biomarkers and therapeutic targets in cancer.
Tumor progression is significantly attributable to a distinctive and suitable tumor microenvironment (TME), that is largely maintained by a variety of stromal cells including cancer stem cells (CSCs), cancer endothelial cells (CECs), cancer-associated fibroblasts (CAFs) and infiltrated immune cells (18, 19). Stromal cells are recruited and activated, acting together to trigger downstream signals that promote tumor formation, angiogenesis and metastasis (20, 21). This has attracted increasing attention aimed at identifying these stromal cells as potential targets for novel cancer therapies. Recently, numerous studies revealed that various lncRNAs play significant roles in the regulation of TME, particularly stromal cells. The main objective of this review is to discuss the basic properties and functional roles of the lncRNAs in the contribution of the TME, to lay a foundation for lncRNAs-based therapies in cancer treatment.
LncRNAs play crucial roles in the modulation of the TME
LncRNAs as modulators of infiltrated immune cells
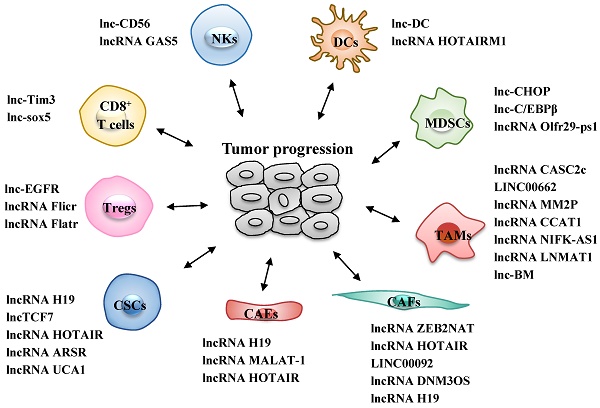
Different types of infiltrated immune cells are important components of the TME and act together to help cancer cells to escape immune surveillance, thus generating a tumor-promoting microenvironment for proliferation and metastasis of cancer cells (22). The roles of lncRNAs in the differentiation and function of various immune cells, including T cells, dendritic cells (DCs), natural killer cells (NKs), tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs), are increasingly well understood (Fig. 1 and Table 1).
LncRNAs act as modulators between immune cells and tumor cells. Immune cells include CD8+ T cells, regulatory T cells (Tregs), dendritic cells (DCs), natural killer cells (NKs), tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs). +: promoting the tumor progression; -: inhibiting the tumor progression; NR: Not Reported.
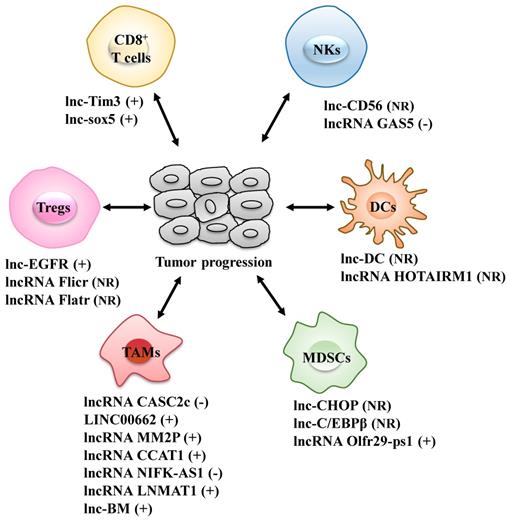
LncRNAs act as modulators between immune cells and tumor cells
| LncRNA | Cancer type | Stromal cells | Mechanism of action | Ref. |
|---|---|---|---|---|
| lnc-EGFR | Hepatocellular carcinoma | Tregs | Lnc-EGFR binds to EGFR, and activates AP-1/NF-AT1/FOXP3 axis, promoting Tregs differentiation and tumor progression | 27 |
| Flicr | - | Tregs | Flicr inhibits the expression of FOXP3 and curtails the immunosuppressive function of Tregs | 28 |
| Flatr | - | Tregs | Flatr promotes the expression of FOXP3 and enhances the immunosuppressive function of Tregs | 29 |
| lnc-Tim3 | Hepatocellular carcinoma | CD8+ T | Lnc-Tim interacts with Tim-3 to release Bat3 and induces exhaustion of CD8+ T cells, exerting HCC immune evasion | 31 |
| lnc-sox5 | Colorectal cancer | CD8+ T | Lnc-sox5 suppresses the infiltration and the cytotoxicity of CD8+ T by increasing IDO1 expression and therefore promotes the tumorigenesis | 8 |
| lnc-DC | - | DCs | Lnc-DC binds directly to the C terminus of STAT3 and activates it, thus promoting DCs differentiation. | 35 |
| HOTAIRM1 | - | DCs | HOTAIRM1 induces upregulation of CD14 and B7H2, thus blocking cells to enter into the DC differentiation pathway | 36 |
| lnc-CD56 | - | NKs | Lnc-CD56 may function as a positive regulator of CD56, and be essential for the developmental and diverse functions of NKs | 39 |
| GAS5 | Liver cancer | NKs | GAS5 regulates miR-544/RUNX3 to enhance the killing effects of NKs, leading to inhibition of tumor growth. | 40 |
| CASC2c | Glioblastoma multiforme | TAMs | CASC2c suppresses the M2 polarization by repressing expression of FX in GBM cells and inhibiting ERK1/2 and AKT in macrophages. | 43 |
| LINC00662 | Hepatocellular carcinoma | TAMs | LINC00662 activates Wnt/β-catenin signaling and further promotes M2 polarization, promoting tumor growth and metastasis. | 44 |
| MM2P | Osteosarcoma | TAMs | MM2P promotes M2 polarization by inducing phosphorylation on STAT6, resulting in macrophage-promoted tumor progression. | 45 |
| CCAT1 | Prostate cancer | TAMs | CCAT1/miR-148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. | 46 |
| NIFK-AS1 | Endometrial cancer | TAMs | NIFK-AS1 inhibits M2 polarization of macrophages and malignant phenotype of endometrial cancer cells through targeting miR-146a. | 47 |
| LNMAT1 | Bladder cancer | TAMs | LNMAT1 activates CCL2 expression to recruit macrophages into the tumor and ultimately promote lymphatic metastasis of bladder cancer. | 50 |
| lnc-BM | Breast cancer | TAMs | lnc-BM induces STAT3-dependent expression of CCL2 to attract macrophages, promoting breast cancer brain metastasis. | 51 |
| lnc-CHOP | - | MDSCs | Lnc-CHOP upregulates the expression of ARG1 and NOS2 to enhance the immunosuppressive function of MDSCs. | 54 |
| lnc-C/EBPβ | - | MDSCs | lnc-C/EBPβ inhibits the expressions of ARG1 and NOS2, to suppress immune-suppressive function and differentiation of MDSCs. | 55 |
| Olfr29-ps1 | Melanoma | MDSCs | Olfr29-ps1promotes functions of monocytic MDSCs and thus tumor growth, through a m6A-modified regulatory network. | 56 |
Tregs: regulatory T cells; DCs: dendritic cells; NKs: natural killer cells; TAMs: tumor-associated macrophages; MDSCs: myeloid-derived suppressor cells.
LncRNAs as modulators of T cells
T cells, a predominant immune cell type in the TME, exert dual roles in tumor progression (23). Cancer cells exploit the immunosuppressive properties of T cells, while weakening the effective functions of anti-tumor T cells (24). LncRNAs have been recognized as important regulators of several T cell functions. Regulatory T cells (Tregs), an immunosuppressive subset of CD4+ T cells characterized by the expression of the master transcription factor forkhead box protein P3 (FOXP3), frequently accumulate in the TME and even represent the major population of infiltrating CD4+ T cells (25, 26). There is growing evidence that some lncRNAs, such as lnc-EGFR, Flicr and Flatr, are involved in Treg biology. Among them, lnc-EGFR is highly expressed in Tregs of patients with hepatocellular carcinoma (HCC), where it acts as an activator of Tregs differentiation. Mechanistically, lnc-EGFR binds specifically to EGFR, inhibits its ubiquitination and subsequent degradation, and sustains the activation of its downstream AP-1 and NF-AT1, two transcription factors for FOXP3, therefore leading to the enhancement of Tregs immunosuppressive function and promotion of HCC progression (27). Both Flicr and Flatr, two lncRNAs conserved and enriched in activated Tregs, were reported to play crucial roles in the regulation of FOXP3 expression and immunosuppressive function of Tregs (28, 29), but their roles in the TME remain unclear. These findings indicate that targeting specific lncRNAs in Tregs has a broad application prospect in the development of anti-tumor therapeutic strategies.
CD8+ T cells, major population of T cells in the TME, exert an efficient anti-tumor attack (30). LncRNAs such as lnc-Tim3 and lnc-sox5 participate in modulating the function of CD8+ T cells. The expression of lnc-Tim3 is up-regulated in HCC patients, which is negatively correlated with the production of IFN-γ and IL-2 by tumor-infiltrating CD8+ T cells. Mechanistically, lnc-Tim3 interacts with Tim-3 to release Bat3, and thereby suppresses downstream Lck/NF-AT1/AP-1 signaling, resulting in the exhaustion of CD8+ T cells and HCC immune evasion (31). Similarly, lnc-sox5 is significantly increased in colorectal cancer (CRC) and correlated with CRC progression. Extensively, lnc-sox5 knock-down dramatically promotes the infiltration and the cytotoxicity of CD8+ T by suppressing the expression of indoleamine 2,3-dioxygenase 1 (IDO1) and therefore suppresses the tumorigenesis of CRC (8). Collectively, these data suggest that dysregulations of lncRNAs in T cells affect immune evasion and tumor progression, and might be potential targets of tumor immunotherapy.
LncRNAs as modulators of dendritic cells
Dendritic cells (DCs) in the TME exhibit an important role in cross-priming CD8+ T cells, in response to initiate and sustain anti-tumor T cells immunity (32-34). Notably, lncRNAs, such as lnc-DC and HOTAIRM1, have been implicated in the DCs differentiation. Wang et al. (35) showed that lnc-DC is exclusively expressed in conventional dendritic cells and driven by the transcription factor PU.1, a key regulator of DCs differentiation. Knockdown of lnc-DC down-regulates the expression of function-related genes and antigens uptake by DCs, thus impairing DCs differentiation from human monocytes and reducing their capacity to stimulate T cell activation. Mechanistic evidence has shown that lnc-DC exerts these effects by activating STAT3, a transcription factor that regulates DCs differentiation. Lnc-DC binds directly to the C terminus of STAT3 to block SHP1-mediated dephosphorylation of STAT3, thus promoting the phosphorylation of STAT3 on tyrosine-705 and the expression of genes associated with DCs activation. Subsequently, Xin et al. (36) reported that lncRNA HOTAIRM1 is downregulated during the process of monocyte differentiating into DCs. HOTAIRM1 competitively binds to miR-3960, hinders miR-3960 from repressing monocyte-related HOXA1 mRNA expression and induces subsequent upregulation of two monocyte markers CD14 and B7H2, thereby sustaining monocyte phenotype and blocking cells entry into the DCs differentiation pathway. However, the role during tumor progression of these lncRNAs within DCs awaits further investigation.
LncRNAs as modulators of natural killer cells
Natural killer cells (NKs) play a critical role in the anti-tumor immune response and participate in controlling tumor progression and metastasis, due to their cytotoxic potential and ability to release immunoregulatory cytokines (37, 38). The typically studied lncRNA in the NKs is lnc-CD56, which is involved in the regulation of CD56, a classical human NKs surface marker. Knockdown of lnc-CD56 reduces the expression of CD56, suggesting lnc-CD56 may be a positive regulator of CD56 and essential for the development and diverse functions of NKs (39). Recently, Fang et al. (40) found that lncRNA GAS5 is down-regulated in NKs of patients with liver cancer, while up-regulated in activated NKs compared with non-stimulated NKs. Overexpression of GAS5 in activated NKs increases IFN-γ secretion, NKs cytotoxicity, and the percentage of CD107a+ NKs through regulating miR-544/RUNX3, hence, enhancing the killing effects of NKs and inhibiting tumor growth. Moreover, down-regulation of GAS5 is associated with liver cancer progression, conferring a worse overall patient survival (41). These findings highlight the importance of lncRNAs in NKs functions and anti-tumor immune response.
LncRNAs as modulators of tumor-associated macrophages
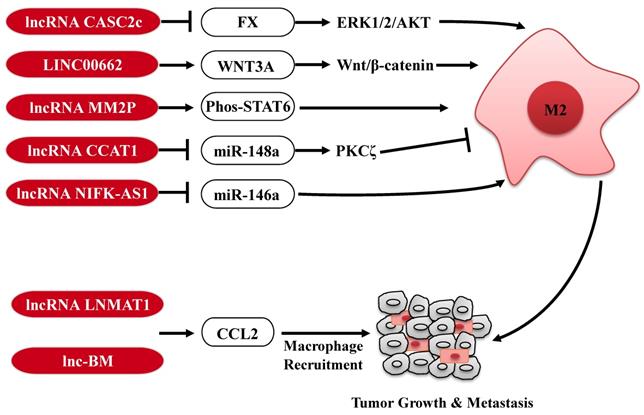
Tumor-associated macrophages (TAMs) are key regulators of the TME, and orchestrate various aspects of tumor progression. In response to microenvironmental signals, TAMs undergo the polarization of pro-inflammatory M1 or anti-inflammatory M2 and, therefore, have anti-tumor or pro-tumor abilities (42). Several lncRNAs expressed in tumor cells are involved in the polarization of TAMs to affect tumor progression (Fig. 2). For example, in glioblastoma multiforme, lncRNA CASC2c inhibits macrophage migration and polarization to the M2 subtype, via binding to coagulation factor X (FX) and commonly inhibiting its expression and secretion. The reduction of FX secreted in the tumor microenvironment results in the decrease of the phosphorylation and activation of ERK1/2 and AKT in macrophages, which plays a crucial role in the M2 macrophage polarization (43). In contrast, in HCC, lncRNA LINC00662 induces WNT3A expression and secretion, and consequently activates Wnt/β-catenin signaling in macrophages in a paracrine manner. Therefore, LINC00662 promotes M2 macrophage polarization and consequently results in HCC tumor growth and metastasis. Clinical data further confirm that high expression of LINC00662 in HCC is correlated with overactivated WNT3A, M2 macrophage polarization and poor prognosis of HCC patients (44). In addition, a high-throughput profiling shows that lncRNA-MM2P is specifically upregulated during the polarization of M2 macrophages. Knockdown of lncRNA-MM2P in macrophages reduces phosphorylation on STAT6, suppresses the transcription of M2-related genes in macrophages, and ultimately blocks M2 macrophage polarization. Thus, targeting lncRNA-MM2P impairs macrophage-promoted tumor angiogenesis and progression (45). Besides, lncRNAs, CCAT1 and NIFK-AS1 expressed in macrophages, also play a key role in modulating the polarization of TAMs in prostate cancer and endometrial cancer, respectively (46, 47).
Furthermore, tumor-derived CCL2 is released into the TME and recruits macrophages to promote tumor progression (48, 49). LncRNAs are also involved in regulation of TAMs infiltration in tumor through affecting CCL2 expression (Fig. 2). For example, lncRNA LNMAT1 is markedly upregulated in lymph node (LN)-metastatic bladder cancer and associated with LN metastasis and prognosis. Specifically, LNMAT1 epigenetically activates CCL2 transcription by enhancing hnRNPL-mediated histone H3 lysine 4 trimethylation (H3K4me3) at the CCL2 promoter. LNMAT1-induced CCL2 in bladder cancer cells contributes to recruit macrophages into the tumor and ultimately promotes lymphatic metastasis of bladder cancer (50). Similarly, high expression of lnc-BM also promotes breast cancer brain metastasis, through inducing STAT3-dependent expression of CCL2 to attract macrophages to the tumor (51). Taken together, lncRNAs expressed in TAMs or secreted by tumor cells modulate the function of TAMs through diverse mechanisms, further affecting tumorigenesis and metastasis, reminding us that targeting these lncRNAs in the TAMs or tumor cells may be a potential anti-tumor strategy.
LncRNAs as modulators of myeloid-derived suppressor cells
Myeloid-derived suppressor cells (MDSCs) are the central cell population with potent immunosuppressive activity on T cells (52, 53). It has been demonstrated that some lncRNAs such as lnc-CHOP and lnc-C/EBPβ, are involved in the modulation of generation, recruitment and immunosuppressive functions of MDSCs. Lnc-CHOP interacts with CHOP and C/EBPβ isoform liver-enriched inhibitory protein (LIP) to promote the activation of C/EBPβ, and results in the expression of major molecules linked to MDSC immunosuppressive activity, including ARG1 and NOS2 (54). In contrast, lnc-C/EBPβ was reported to bind with LIP to inhibit the activation of C/EBPβ, and further reduce immunosuppressive function and differentiation of MDSCs (55). A recent study showed that lncRNA Olfr29-ps1, is expressed in MDSCs and upregulated by the proinflammatory cytokine IL6. Olfr29-ps1 promotes immunosuppressive function and differentiation of monocytic MDSCs, through a N6-methyladenosine (m6A)-modified regulatory network (56). These findings reveal diverse mechanisms by which lncRNAs regulate the function of MDSCs and also provide potential therapeutic targets.
LncRNA as modulators of cancer stem cells
Cancer stem cells (CSCs), a rare sub-population within tumor, are key components of the TME, that have the ability of self-renewal and limitless proliferation (57). CSCs are believed to be responsible for tumor initiation, progression and resistance to therapies (58, 59). Several studies described that various lncRNAs are able to modulate self-renewal, maintenance and differentiation of CSCs through different molecular mechanisms (Fig. 3 and Table 2). To date, lncH19, lncTCF7, lncARSR, UCA1 and HOTAIR are the most highlighted lncRNAs in CSCs.
The regulatory roles of lncRNAs in TAMs. LncRNAs are involved in regulation the polarization of TAMs or CCL2-mediated macrophages recruitment to affect tumor progression. M2: M2 macrophage.
LncRNAs act as modulators between cancer stem cells (CSCs)/cancer-associated endothelial cells (CAEs)/cancer-associated fibroblasts (CAFs) and tumor cells
| LncRNA | Cancer type | Stromal cells | Mechanism of action | Ref. |
|---|---|---|---|---|
| H19 | Prostate cancer | CSCs | H19 increases the expression of Oct4 and Sox2 to promote the sphere-forming capacity. | 61 |
| H19 | Breast cancer | CSCs | H19 sponges and inhibits miRNA let-7 and enhances the expression of LIN28, contributing to the maintenance of stem cells. | 62 |
| lncTCF7 | Hepatocellular carcinoma | CSCs | lncTCF7 activates the Wnt signaling cascade to prime CSC self-renewal and tumor propagation. | 63 |
| HOTAIR | Liver cancer | CSCs | HOTAIR enhances liver CSC growth through dissociating the CREB-p300-RNApolII complex to repress expression of SETD2. | 68 |
| HOTAIR | Breast cancer | CSCs | HOTAIR maintains BCSCs self-renewal capacity by negatively regulating miR-34a and consequently Sox2. | 65 |
| ARSR | Renal carcinoma | CSCs | ARSR promotes the expansion of renal CSCs through interaction with YAP, to block its phosphorylation. | 69-71 |
| UCA1 | Liver cancer | CSCs | UCA1 promotes the malignant transformation of hepatocyte-like stem cells via activating telomere length extension and c-Myc expression. | 78 |
| H19 | Glioma | CAEs | H19 promotes glioma-induced endothelial cell proliferation, migration and tube formation via increasing the expression of VASH2. | 82 |
| H19 | Liver cancer | CAEs | CD90+ liver cancer cells package H19 inside exosomes, which is released to promote functions of endothelial cells. | 83 |
| MALAT-1 | - | CAEs | MALAT-1 promotes the proliferation rate of HUVECs by upregulation of FOMX1 expression. | 84 |
| MALAT-1 | Neuroblastoma | CAEs | MALAT1 promotes endothelial cell migration, invasion and vasculature formation via increasing the expression of FGF2. | 85,86 |
| HOTAIR | Nasopharyngeal carcinoma | CAEs | HOTAIR promotes endothelial cell tube formation and angiogenesis through activating the transcription of VEGFA and Ang2 expression. | 87 |
| HOTAIR | Glioma | CAEs | HOTAIR promotes the angiogenic function by activating VEGFA and glioma cell-derived vesicles. | 88 |
| ZEB2NAT | Bladder cancer | CAFs | ZEB2NAT promotes the CAFs-mediated initiation of efficient metastasis in a TGFβ1-dependent process. | 93 |
| HOTAIR | Breast cancer | CAFs | HOTAIR promotes the CAFs-mediated tumor development in a TGFβ1-dependent process. | 94 |
| LINC00092 | Ovarian cancer | CAFs | CAFs-secrete CXCL14 induces LINC00092 upregulation to promote tumor metastasis by enhancing PFKFB-2 translation. | 95 |
| DNM3OS | Esophageal cancer | CAFs | CAFs-promoted expression of DNM3OS confers significant radio-resistance via regulating DNA damage response. | 96 |
| H19 | Colorectal cancer | CAFs | H19 expressed by CAFs contributes to chemo-resistance of colorectal cancer through activating the β-catenin pathway. | 97 |
lncRNA H19 is one of the first non-coding RNAs identified as a cancer-related lncRNA (60). In prostate cancer, H19 level modulations are positively correlated with the expression of key pluripotency transcription factors (Oct4, Sox2) and the sphere-forming capacity, uncovering a role for H19 as a potential stemness regulator (61). In breast cancer, H19 acts to sponge miRNA let-7 and inhibit its biological function, subsequently leading to the elevation of LIN28, the core pluripotency factor that is crucial for the maintenance of breast cancer stem cells (BCSCs) (62). In HCC, lncTCF7 recruits the SWI/SNF chromatin remodeling complex to TCF7 promoter to regulate TCF7 transcription, activating the Wnt signaling cascade and thus priming liver CSCs self-renewal and tumor propagation (63).
Hox transcript antisense intergenic RNA (HOTAIR) is an oncogenic lncRNA, the expression of which is elevated in multiple CSCs (64-66), and positively associated with advanced tumor progression and poor prognosis (67). HOTAIR is able to promote liver cancer stem cell malignant growth through downregulation of SETD2, a specific methyltransferase for histone H3 lysine 36 (H3K36me3) and required for ATM activation upon DNA double-strand breaks (DSBs) (68). Additionally, HOTAIR is highly up-regulated in BCSCs models, and tightly regulates self-renewal capacity of CSCs through transcriptional inhibition of miR-34a and consequent upregulation of Sox2 (65).
The lncRNA activated in renal cell carcinoma with sunitinib resistance (ARSR) was recently identified as a novel lncRNA. ARSR is highly expressed in primary renal CSCs and predicts poor prognosis. ARSR processes self-renewal capacity and promotes the metastasis of renal CSCs. Mechanistically, ARSR interacts with Yes-associated protein (YAP) to block its phosphorylation by LATS1, thus facilitating YAP nuclear translocation, which is required to sustain CSCs self-renewal (69-71).
The lncRNA urothelial cancer associated 1 (UCA1) is highly expressed in multiple human cancers, including hepatocellular cancer, gastric cancer, colorectal cancer and lung cancer, which confers a worse overall patient survival (72-77). Notably, UCA1 plays a critical role in governing growth and malignant transformation of CSCs through the upregulation and activation of telomerase reverse transcriptase (TERT) and oncogene C-myc. Mechanistically, excessive UCA1 leads to increased binding capacity of UCA1 to CyclinD1. Therefore, UCA1-CyclinD1 complex is recruited to c-Myc promoter region, increasing the outcome of oncogene C-myc (78). On the other hand, UCA1-CyclinD1 complex also activates lncRNA H19 transcription via reducing DNA methylation on H19 promoter region. Strikingly, overexpression of H19 enhances the binding of TERT, thus enhancing the cell telomerase activity and extending the telomere length (78). Besides, UCA1 enhances the phosphorylation of RB1, which could promote the interplay between histone lysine methyltransferase SET1A and pRB1. Then, the complex induces the trimethylation of H3K4 on telomere capping essential gene TRF2 promoter region, causing TRF2 overexpression and consequently prolonging the telomere length (79). Taken together, these observations highlight the critical role of multiple lncRNAs in modulating CSCs maintenance and self-renewal and provide a potential application for targeting lncRNAs as an alternative effective strategy.
LncRNAs as modulators of cancer-associated endothelial cells
Cancer-associated endothelial cells (CAEs), important components of the TME, are responsible for angiogenesis and regulation of tumor growth and metastasis (80, 81). Altered expression of lncRNAs in CAEs also affects tumor progression through modulating the biological behaviors of CAEs (Fig. 3 and Table 2).
H19 expression is significantly up-regulated in glioma microvessels and glioma-induced endothelial cells (GECs). H19 regulates the proliferation, migration and tube formation of GECs by targeting miR-29a, which decreases the expression of vasohibin 2 (VASH2), an angiogenic factor (82). H19 derived from tumor cells could also affect angiogenesis of CAEs. CD90+ liver cancer cells package H19 inside exosomes, which is released to influence endothelial cells by promoting angiogenesis and stimulating their adhesive properties (83).
MALAT1 can also regulate the properties of CAEs. Researchers have found that knockdown of MALAT1 expression significantly inhibits the proliferation of human umbilical vein endothelial cells (HUVECs), which is mediated by upregulation of miR-320a and hence, downregulation of cell cycle regulator FOMX1 expression in HUVECs (84). In neuroblastoma, up-regulation of MALAT1, induced by hypoxia, results in endothelial cell migration, invasion and vasculature formation via increasing the expression of the pro-angiogenic factor, FGF2 (85, 86). These data demonstrate that MALAT1 plays an important role in endothelial cell proliferation and tumor angiogenesis.
LncRNAs act as modulators between cancer stem cells (CSCs)/cancer-associated endothelial cells (CAEs)/cancer-associated fibroblasts (CAFs) and tumor cells. +: promoting the tumor progression.
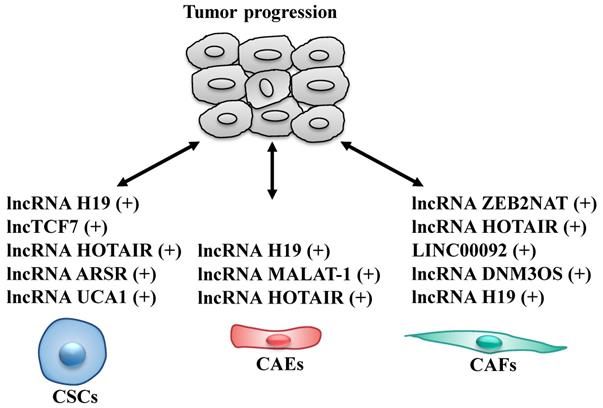
HOTAIR is extremely abundant in nasopharyngeal carcinoma cells (NPC) and functions as an angiogenesis activator. Specifically, HOTAIR promotes endothelial cell tube formation and angiogenesis through directly activating the transcription of angiogenic factor VEGFA (87). Furthermore, in glioma cells, the angiogenic function of HOTAIR is mediated not only by the regulation of VEGFA expression, but also by direct transmission into endothelial cells via glioma cell-derived vesicles (88). Collectively, deregulated lncRNAs expressions in CAEs do affect the tumor progression, suggesting that targeting lncRNAs both in CAEs and tumor cells might be a new approach for cancer therapy.
LncRNA as modulators of cancer-associated fibroblasts
Cancer-associated fibroblasts (CAFs), the activated fibroblasts, are one of the most dominant components of the tumor microenvironment (89, 90). CAFs are able to promote tumor progression, such as proliferation, invasion and angiogenesis, through the secretion of growth factors, cytokines and chemokines (91). Although the role of lncRNA in CAFs modulation is poorly investigated, some studies suggested that they can contribute to: i) CAFs phenotype and function; ii) enhancement of CAFs-triggering signals (e.g. TGF-β1) to induce epithelial-mesenchymal transition (EMT) and metastasis of tumor cells (Fig. 3 and Table 2). High-throughput sequencing technologies reveal numerous novel lncRNAs differentially expressed in distinguishing CAFs from normal ovarian fibroblasts (NOFs), followed by the functional network to predict those specific lncRNAs involved in the pro-metastatic phenotype of CAFs (92).
TGF-β1 secreted by CAFs induces the metastatic activity of cancer cells by regulating the expression of lncRNAs. For example, CAFs-mediated upregulation of lncRNA ZEB2NAT transcription in bladder cancer cells that promotes EMT via the secretion of TGF-β1, which is linked to poor clinical outcome (93). Similarly, CAFs-secreted TGF-β1 induces the transcription of lncRNA HOTAIR to promote EMT and metastasis in breast cancer cells (94). Moreover, the lncRNA, LINC00092, is induced upon stimulation by CAF-secreted CXCL14 in ovarian cancer and correlated with poor prognosis in patients. Mechanistically, LINC00092 binds with fructose-2, 6-biphosphatase 2 (PFKFB2), thereby promoting ovarian cancer metastasis by altering glycolysis and sustaining CAFs-like features of fibroblasts (95).
In addition, CAF-promoted lncRNAs are also involved in radio-resistance and chemo-resistance. In esophageal cancer cells, CAFs-promoted expression of lncRNA DNM3OS confers significant radio-resistance via regulating DNA damage response in a FOXO1-dependent manner (96). Moreover, lncRNA H19 expressed by CAFs was reported to contribute to chemo-resistance of colorectal cancer through activating the β-catenin pathway (97). Collectively, these studies indicate the importance of lncRNAs in the interaction between the CAFs and cancer cells, reminding us that targeting lncRNAs could be a new approach for cancer therapy.
Conclusion and further perspective
Here, we have summarized recent advancement involving of lncRNAs within the tumor microenvironment and their roles in the crosstalk between infiltrated immune cells, CSCs, CAEs, CAFs and tumor cells, as well as some of the underlying molecular mechanisms. lncRNAs exert their functions in different ways to modulate tumor growth and progression. As yet, only a few lncRNAs have been well-studied in the tumor-stroma crosstalk, warranting further studies on the identification of more new types of lncRNAs and their mechanisms involved in the future. A better understanding of the role of lncRNAs within the tumor microenvironment may lead to the discovery of potential biomarkers and development of novel targeted therapies.
At present, the pressing issue is to systematically elucidate the key aspects of lncRNAs, including the expression, structure, function and regulatory mechanism. The improvement of analytical technologies for the specific biological functions of lncRNAs may help explore its greatest relevance to various cancers. Such information will provide a basis for considering lncRNAs as ideal diagnostic markers or even therapeutic targets. In addition, approaches of targeting lncRNAs should be considered and optimized, such as the use of siRNA to induce lncRNA degradation and CRISPR/Cas9 mediated gene editing. How to specifically deliver the respective molecules into targeted cells is still a great challenge.
In summary, the functional importance of lncRNAs within tumor microenvironment is gradually characterized; clinical application of lncRNAs still needs to be studied further. With the deep-going research, lncRNAs-associated tumor-stroma crosstalk will open up a new era of anti-tumor therapy.
Abbreviations
AKT: AKT serine/threonine kinase 1; ARG1: arginase-1; ARSR: activated in renal cell carcinoma with sunitinib resistance; BCSC: breast cancer stem cell; C/EBPβ: CCAAT enhancer binding protein beta; CAE: cancer-associated endothelial cell; CAF: cancer-associated fibroblast; CASC2c: CASC2 cancer susceptibility 2; CCAT-1: colon cancer‐associated transcript‐1; CCL2: C-C motif chemokine ligand 2; CHOP: C/EBP homologous protein; CRC: colorectal cancer; CSC: cancer stem cell; CXCL14: C-X-C motif chemokine ligand 14; DC: dendritic ell; DSB: DNA double-strand break; EGFR: epidermal growth factor receptor; EMT: epithelial-mesenchymal transition; ERK1/2: extracellular signal-related kinase 1/2; FGF2: fibroblast growth factor 2; Flatr: Foxp3-specific lncRNA anticipatory of Tregs; Flicr: FOXP3 regulating long intergenic non-coding RNA; FOMX1: forkhead box M1; FOXO1: forkhead box O1; FOXP3: forkhead box protein P3; GAS5: growth arrest specific 5; GEC: glioma-induced endothelial cells; HCC: hepatocellular carcinoma; hnRNPL: heterogeneous nuclear ribonucleoprotein L; HOTAIR: HOX transcript antisense RNA; HOTAIRM1: HOXA transcript antisense RNA, myeloid-specific 1; HOXA1: homeobox A1; HUVEC: human umbilical vein endothelial cell; IDO1: indoleamine 2,3-dioxygenase 1; IFN-γ: interferon gamma; IL-2: interleukin 2; IL-6: interleukin 6; LATST1: large tumor suppressor kinase 1; LIN28: lin-28 homolog A; LIP: liver-enriched inhibitory protein; lncRNAs: long noncoding RNAs; LNMAT1: lymph node metastasis associated transcript 1; MALAT1: metastasis associated lung adenocarcinoma transcript 1; MDSC: myeloid-derived suppressor cell; NIFK-AS1: NIFK antisense RNA 1; NK: natural killer; NOS2: NO synthase 2; NPC: nasopharyngeal carcinoma cell; PCA3: prostate cancer associated 3; PFKFB2: fructose-2, 6-biphosphatase 2; RB1: RB transcriptional corepressor 1; RCC: renal cell carcinoma; RUNX3: RUNX family transcription factor 3; SET1A: SET domain containing 1A, histone lysine methyltransferase; SETD2: SET domain containing 2, histone lysine methyltransferase; STAT3: signal transducer and activator of transcription 3; STAT6: signal transducer and activator of transcription 6; TAM: tumor-associated macrophage; TERT: telomerase reverse transcriptase; TGF-β: transforming growth factor beta; TME: tumor microenvironment; Treg: regulatory T cell; TRF2: TATA box binding protein-related factor 2; UCA1: urothelial cancer associated 1; VASH2: vasohibin 2; VEGFA: vascular endothelial growth factor A; WNT3A: Wnt family member 3A; YAP: Yes-associated protein.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81871870 and 81702244), Zhejiang Provincial Medical Association Project (Grant Nos. 2013zyc-a138), and Science and Technology Bureau of Jiaojiang district (Grant Nos. 132055).
Author Contributions
LSZ and YYZ drafted, DSS and QZ revised and finalized the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Iyer MK, Niknafs YS, Malik R. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199-208
2. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178-81
3. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904-14
4. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253-61
5. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452-63
6. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391-407
7. Zhu M, Chen Q, Liu X. et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766-75
8. Wu K, Zhao Z, Liu K. et al. Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell Cycle. 2017;16(13):1295-301
9. Zheng X, Zhang Y, Liu Y. et al. HIF-2alpha activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem. 2018;119(4):3247-56
10. de Kok JB, Verhaegh GW, Roelofs RW. et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62(9):2695-8
11. Hessels D, Klein Gunnewiek JM, van Oort I. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44(1):8-15 discussion -6
12. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7(5):582-5
13. Raveh E, Matouk IJ, Gilon M. et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184
14. Ayesh S, Matouk I, Schneider T. et al. Possible physiological role of H19 RNA. Mol Carcinog. 2002;35(2):63-74
15. Lavie O, Edelman D, Levy T. et al. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch Gynecol Obstet. 2017;295(3):751-61
16. Gofrit ON, Benjamin S, Halachmi S. et al. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. 2014;191(6):1697-702
17. Hanna N, Ohana P, Konikoff FM. et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. 2012;19(6):374-81
18. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423-37
19. Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51(1):27-41
20. Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309-22
21. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-74
22. Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715-27
23. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32(19-20):1267-84
24. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883-99
25. Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16(10):599-611
26. Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol. 2014;27:1-7
27. Jiang R, Tang J, Chen Y. et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129
28. Zemmour D, Pratama A, Loughhead SM. et al. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci U S A. 2017;114(17):E3472-E80
29. Brajic A, Franckaert D, Burton O. et al. The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Front Immunol. 2018;9:1989
30. Hanson HL, Donermeyer DL, Ikeda H. et al. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity. 2000;13(2):265-76
31. Ji J, Yin Y, Ju H. et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018;9(5):478
32. Gardner A, Ruffell B. Dendritic Cells and Cancer Immunity. Trends Immunol. 2016;37(12):855-65
33. Kurts C, Robinson BW, Knolle PA. Cross-priming in health and disease. Nat Rev Immunol. 2010;10(6):403-14
34. Fu C, Jiang A. Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment. Front Immunol. 2018;9:3059
35. Wang P, Xue Y, Han Y. et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310-3
36. Xin J, Li J, Feng Y. et al. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. Onco Targets Ther. 2017;10:1307-15
37. Terme M, Ullrich E, Delahaye NF. et al. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nat Immunol. 2008;9(5):486-94
38. Smyth MJ, Hayakawa Y, Takeda K. et al. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850-61
39. Zhang R, Ni F, Fu B. et al. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget. 2016;7(45):72546-58
40. Fang P, Xiang L, Chen W. et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019;25(2):99-109
41. Chang L, Li C, Lan T. et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13(2):1541-50
42. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953-64
43. Zhang Y, Feng J, Fu H. et al. Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front Immunol. 2018;9:1557
44. Tian X, Wu Y, Yang Y. et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/beta-catenin signaling. Mol Oncol. 2019
45. Cao J, Dong R, Jiang L. et al. LncRNA-MM2P Identified as a Modulator of Macrophage M2 Polarization. Cancer Immunol Res. 2019;7(2):292-305
46. Liu J, Ding D, Jiang Z. et al. Long non-coding RNA CCAT1/miR-148a/PKCzeta prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79(1):105-12
47. Zhou YX, Zhao W, Mao LW. et al. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int J Biochem Cell Biol. 2018;104:25-33
48. Bonapace L, Coissieux MM, Wyckoff J. et al. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 2014;515(7525):130-3
49. Pena CG, Nakada Y, Saatcioglu HD. et al. LKB1 loss promotes endometrial cancer progression via CCL2-dependent macrophage recruitment. J Clin Invest. 2015;125(11):4063-76
50. Chen C, He W, Huang J. et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9(1):3826
51. Wang S, Liang K, Hu Q. et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. 2017;127(12):4498-515
52. Kumar V, Patel S, Tcyganov E. et al. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016;37(3):208-20
53. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162-74
54. Gao Y, Wang T, Li Y. et al. Lnc-chop Promotes Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor and Inflammatory Environments. J Immunol. 2018;200(8):2603-14
55. Gao Y, Sun W, Shang W. et al. Lnc-C/EBPbeta Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol Res. 2018;6(11):1352-63
56. Shang W, Gao Y, Tang Z. et al. The Pseudogene Olfr29-ps1 Promotes the Suppressive Function and Differentiation of Monocytic MDSCs. Cancer Immunol Res. 2019;7(5):813-27
57. Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111-5
58. Adorno-Cruz V, Kibria G, Liu X. et al. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75(6):924-9
59. D'Andrea V, Panarese A, Tonda M. et al. Cancer stem cells as functional biomarkers. Cancer Biomark. 2017;20(3):231-4
60. Brannan CI, Dees EC, Ingram RS. et al. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28-36
61. Bauderlique-Le Roy H, Vennin C, Brocqueville G. et al. Enrichment of Human Stem-Like Prostate Cells with s-SHIP Promoter Activity Uncovers a Role in Stemness for the Long Noncoding RNA H19. Stem Cells Dev. 2015;24(10):1252-62
62. Peng F, Li TT, Wang KL. et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8(1):e2569
63. Wang Y, He L, Du Y. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413-25
64. Dou J, Ni Y, He X. et al. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016;8(1):98-108
65. Deng J, Yang M, Jiang R. et al. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem-Like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS One. 2017;12(1):e0170860
66. Padua Alves C, Fonseca AS, Muys BR. et al. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31(12):2827-32
67. Min SN, Wei T, Wang XT. et al. Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: A meta-analysis. Medicine (Baltimore). 2017;96(23):e7084
68. Li H, An J, Wu M. et al. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6(29):27847-64
69. Qu L, Wu Z, Li Y. et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat Commun. 2016;7:12692
70. Song S, Ajani JA, Honjo S. et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014;74(15):4170-82
71. Basu-Roy U, Bayin NS, Rattanakorn K. et al. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Commun. 2015;6:6411
72. Wang F, Ying HQ, He BS. et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899-917
73. Han Y, Yang YN, Yuan HH. et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46(5):396-401
74. Bian Z, Jin L, Zhang J. et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892
75. Ni B, Yu X, Guo X. et al. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. Int J Oncol. 2015;47(4):1329-38
76. Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(11):7938-44
77. Zheng Q, Wu F, Dai WY. et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17(8):640-6
78. Pu H, Zheng Q, Li H. et al. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget. 2015;6(38):40775-98
79. Li T, Zheng Q, An J. et al. SET1A Cooperates With CUDR to Promote Liver Cancer Growth and Hepatocyte-like Stem Cell Malignant Transformation Epigenetically. Mol Ther. 2016;24(2):261-75
80. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501(7467):346-54
81. Maishi N, Hida K. Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 2017;108(10):1921-6
82. Jia P, Cai H, Liu X. et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359-69
83. Conigliaro A, Costa V, Lo Dico A. et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155
84. Sun JY, Zhao ZW, Li WM. et al. Knockdown of MALAT1 expression inhibits HUVEC proliferation by upregulation of miR-320a and downregulation of FOXM1 expression. Oncotarget. 2017;8(37):61499-509
85. Tee AE, Liu B, Song R. et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget. 2016;7(8):8663-75
86. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721-32
87. Fu WM, Lu YF, Hu BG. et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7(4):4712-23
88. Ma X, Li Z, Li T. et al. Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am J Transl Res. 2017;9(11):5012-21
89. Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99-115
90. Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582-98
91. De Jaeghere EA, Denys HG, De Wever O. Fibroblasts Fuel Immune Escape in the Tumor Microenvironment. Trends Cancer. 2019;5(11):704-23
92. Vafaee F, Colvin EK, Mok SC. et al. Functional prediction of long non-coding RNAs in ovarian cancer-associated fibroblasts indicate a potential role in metastasis. Sci Rep. 2017;7(1):10374
93. Zhuang J, Lu Q, Shen B. et al. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924
94. Ren Y, Jia HH, Xu YQ. et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. 2018;17(1):5
95. Zhao L, Ji G, Le X. et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017;77(6):1369-82
96. Zhang H, Hua Y, Jiang Z. et al. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25(6):1989-2000
97. Ren J, Ding L, Zhang D. et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932-48
Author contact
![]() Corresponding author: Dongsheng Sun: Taizhou University hospital, Taizhou University, Taizhou, Zhejiang, China. Tel: +8618767666287. Email: sundsedu.cn. Qiang Zhang: Taizhou Municipal Hospital, Taizhou University, Taizhou, Zhejiang, China. Tel: +8613806586602. Email: z-q2007com
Corresponding author: Dongsheng Sun: Taizhou University hospital, Taizhou University, Taizhou, Zhejiang, China. Tel: +8618767666287. Email: sundsedu.cn. Qiang Zhang: Taizhou Municipal Hospital, Taizhou University, Taizhou, Zhejiang, China. Tel: +8613806586602. Email: z-q2007com

 Global reach, higher impact
Global reach, higher impact