10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(11):2899-2911. doi:10.7150/ijbs.58127 This issue Cite
Review
Insight for Immunotherapy of HCMV Infection
1. Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha 410013, Hunan, China
2. The Key Laboratory of Carcinogenesis of the Chinese Ministry of Health, The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute, Central South University, Changsha, 410008 , Hunan, China
3. Department of Pathogeny Biology, School of Basic Medical Science, Central South University, Changsha, 410078, Hunan, China
Received 2021-1-16; Accepted 2021-6-30; Published 2021-7-13
Abstract
Human cytomegalovirus (HCMV), a ubiquitous in humans, has a high prevalence rate. Young people are susceptible to HCMV infection in developing countries, while older individuals are more susceptible in developed countries. Most patients have no obvious symptoms from the primary infection. Studies have indicated that the virus has gradually adapted to the host immune system. Therefore, the control of HCMV infection requires strong immune modulation. With the recent advances in immunotherapy, its application to HCMV infections is receiving increasing attention. Here, we discuss the immune response to HCMV infection, the immune escape mechanism, and the different roles that HCMV plays in various types of immunotherapy, including vaccines, adoptive cell therapy, checkpoint blockade therapy, and targeted antibodies.
Keywords: human cytomegalovirus, immunotherapy, immune response, immune escape mechanism, infection
Introduction
Human cytomegalovirus (HCMV), also known as human herpesvirus 5 (HHV-5), is a linear double-stranded DNA β-herpesvirus that belongs to the family of herpesviruses [1]. HCMV infection results in the enlargement of infected tissue cells and the formation of large nuclear inclusion bodies. HCMV was first isolated in 1957 from an infant who was suspected to have congenital toxoplasmosis [2].
A recent systematic review and meta-analysis estimated a global HCMV seroprevalence of 83% in the general population, 86% in women of reproductive age, and 86% in donors of organs or blood [3]. HCMV can be transmitted through mother-to-child transmission; horizontal transmission; hospital-acquired respiratory, neurological, blood, and digestive system diseases; and other multi-system diseases, resulting in fetal malformation, birth defects [4], or poor prognosis of transplant patients with low immunity [5-8]. Following the establishment of infection, HCMV is usually latent (with only a few virus particles being released outside the cell), and the infected persons remain asymptomatic and become lifelong infectious carriers [9]. However, it is a huge health threat for immunocompromised patients, such as newborns [10, 11], AIDS patients [12], and transplant recipients [13]. It often causes infectious mononucleosis-like syndrome, retinitis, pneumonitis, gastrointestinal diseases, mental retardation, and vascular disorders [14]. Interestingly, recent studies have reported the presence of active HCMV infection in gliomas [15] and breast cancer [16]. HCMV infection induces the activation of mitotic signals transmitted by the products of proto-oncogenes such as c-fos c-jun and c-myc, which may be involved the tumorigenicity of HCMV [17].
Currently used drugs, such as Ganciclovir (GCV), Cidofovir (CDV) and Foscavir (FOS), have problems such as poor antiviral effect, large side effects and drug resistance, which cannot meet the clinical needs. Therefore, it is urgent to develop new prevention and treatment methods [18]. Antiviral drugs used in the treatment of patients with HCMV infection have low oral bioavailability and dose-related toxicities, such as bone marrow suppression, hepatotoxicity, and nephrotoxicity [19, 20]. In addition, vaccines against HCMV are being developed, but no licensed vaccine is available thus far [21]. Fortunately, a number of studies have shown that immunotherapy is a promising strategy to overcome the side effects of antiviral treatment and for the development of prevention measures.
The structural characteristics of HCMV
The virion of HCMV is composed of double-stranded DNA (dsDNA), which is enclosed in a capsid comprising four integral protein (pUL46, pUL80.5, pUL85, pUL104) that are organized into 162 capsomeres (150 hexamers plus 12 pentamers) and 320 triplexes located between the capsomeres [22]. The capsid, with a diameter of 100 nm, is surrounded by a poorly defined area and enclosed by a lipid bilayer envelope containing viral glycoproteins providing a final diameter of approximately 180 nm for the whole virion [23]. The HCMV genome is the largest known human herpes virus genome. Recent analysis has suggested that the DNA genome of HCMV consists of 235,000 base pairs, encoding at least 700 open reading frames [24]. The structure of the HCMV genome is similar to that of other herpes virus members. It is composed of the unique sequence of long component (UL) and the unique sequence of short component (US) and has reverse repeating sequences, specifically terminal repeat of long component (TRL), inverted repeat of long component (IRL), inverted repeat of short component (IRS), and terminal repeat of short component (TRS) The order of expression was immediate-early (IE), early (E), and late (L) genes. IE genes are expressed within hours after infection and initiate the transcription of viral genes essential for genome replication. E genes encode proteins, such as DNA polymerase and the terminase complex, which contribute to viral DNA replication and packaging and regulate host cell functions to facilitate virus replication. L genes encode structural proteins of the outer tegument layer and the envelope required for the assembly of the infectious virion [25]. HCMV genome encodes approximately 54 membrane proteins including at least 25 membrane glycoproteins found in the virion envelope [26-28]. A well-defined list of HCMV membrane proteins necessary for virus entry into the cells is not available because most HCMV mutants have been classified as essential or nonessential for replication, and have not been tested for defects in virus entry [29, 30]. Glycoprotein B (gB), glycoprotein H (gH), glycoprotein L (gL), glycoprotein M (gM), and glycoprotein N (gN) are the main HCMV-encoded glycoproteins. gB is a viral fusogene that is essential for entry into all cell types, [31]. Furthermore, gB (UL55) is highly expressed and an immune-dominant target following natural infection, making it an attractive target for vaccination [32]. The gH/gL assemble into UL128, UL130, and UL131 proteins producing gH/gL/UL128-131, which are found in the virion envelope and mediate infection of epithelial and endothelial cells and monocyte-macrophages [33-35]. Among HCMV proteins, the tegument phosphoprotein pp65 (pUL83) is a common clinically detected antigen and plays a major role in immunomodulation and immune evasion.
The life cycle of HCMV
The life cycle of HCMV starts when it binds to receptors on the surface of host cells and enters into the cytoplasm. Current data support two models of HCMV entering the cytoplasm through receptor-mediated endocytosis and membrane fusion [36]. One model involves the interaction of the trimeric gH/gL/gO complex (TC) with platelet-derived growth factor receptor alpha (PDGFRα), activating gB to fuse the virus envelope with the membrane of fibroblasts [37]. The other model involves the interaction of the pentameric gH/gL/UL128-131 complex (PC) with neuropilin-2 (Nrp2), which promotes endocytosis of virus particles in epithelial cells/endothelial cells, followed by gB activation by gH/gL/gO (or gH/gL) and entry into the cytoplasm through endosomes [38]. The network of microtubules (MTs) are closely related to the entry of HCMV into the nucleus. When the MTs are depolymerized by nocodazole, fluorescently labelled pp150 is blocked in the cytoplasm. Furthermore, in the absence of the MT network, the capsids that have entered the cytoplasm do not move in close proximity to the nucleus, suppressing IE gene expression [39, 40]. After the entry of viral DNA into the cell nucleus, cellular RNA polymerases I and II (Pol I and II) are employed to transcribe viral genes by binding to the major immediate early promoter (MIEP) [41]. Previous studies have demonstrated that inhibition of Rho-associated coiled-coil kinase (ROCK) protein 1 results in increased levels of tegument protein UL32 and viral DNA in the cytoplasm, suggesting that ROCK activity hinders efficient egress of HCMV particles out of the nucleus [42]. Such impairments have been previously linked to the failure to control HCMV infection [43]. In addition, The transcriptome of HCMV-infected cells showed features of a pro-oncogenic cellular environment with upregulated expression of multiple oncogenes, enhanced activation of pro-survival genes, cell proliferation, and upregulated markers of stem cells and epithelial mesenchymal transition (EMT) [44].
There are two types of cell infection status: latent infection and lysogenic infection. For a latent infection, latency-associated transcript (LAT) HCMV genes are transcribed and accumulated in host cells [45]. A number of HCMV LATs have been identified, including UL138 [46], latent undefined nuclear antigen (LUNA) [47], UL81-82 [48], US28 [49, 50], and pUL11 [51]. Experimentally, latently infected CD34+ and CD14+ cells have been shown to secrete chemokines, which can recruit T cells, as well as IL-10 and TGF-β, both of which can modulate the activity of T cells that have migrated to the environment surrounding the latent infection [51, 52]. In this case, viruses can indefinitely persist in host cells and have a latent infection pathway. Primary infection may be accompanied by a limited illness, and long-term latency is often asymptomatic [53]. Reactivation of HCMV in IL-6-stimulated dendritic cells (DC) has been reported to be dependent on ERK-MAPK pathway [54].
The immune response of HCMV infection
The reason why we have not found an effective way to combat HCMV is probably that the immune responses to HCMV infections are still poorly understood. Changes in the level of the immune response directly affect the severity of HCMV infection. However, how the immune system responds to HCMV infection is not yet clear.
It is well known that cell-mediated immunity and antibody-mediated immunity are necessary to suppress HCMV infection [55-57].Firstly, HCMV can infect a broad range of cells, including epithelial cells of glandular and mucous tissues, smooth muscle cells, fibroblasts, macrophages [58], hepatocytes, dendritic cells [59], and vascular endothelial cells [60], all of which provide a platform for the efficient proliferation of HCMV [36]. HCMV infection of precursor monocytes leads to the generation of toll-like receptors (TLRs) responsive inflammatory macrophages resistant to down-regulatory stromal TGF-β, allowing the macrophages to react to invading pathogens and immunostimulatory products with inflammation mediated by the Smad7 cytokine response [61].Within the innate immune system, natural killer (NK) cells act as the first line of defense and play an important role in limiting early cytomegalovirus (CMV) infection [62]. Monocytes play a pivotal role in viral dissemination in organ tissues during primary infection and subsequent reactivation from latency [63]. One hallmark of CMV infection is the maintenance of large populations of CMV-specific memory CD8+ T cells, a phenomenon termed memory inflation, and emerging data suggest that memory inflation is associated with impaired immunity in the elderly [55]. HCMV can activate the expression of B cell-activating factor (BAFF-R), thereby promoting polyclonally activated B lymphocytes [64]. HCMV pp65 may be an autoimmune or lupus-prone B cell epitope and may catalyze further epitope spreading to induce anti-dsDNA antibody production in lupus-susceptible individuals [65]. pp65 inhibits interleukin-1β (IL-1β) in an NF-κB-dependent manner, induces IL-1β in a caspase-8 dependent manner [66], and together with IE1 antigen efficiently induces and expands virus-specific T cells [67]. There are still several proteins that play important roles in HCMV infection and immunity, which are worth further exploration and research.
Currently, CD4+ or CD8+ T cells and NK cells are considered to play significant roles in the host defense against HCMV, and they are the key participants in the cellular immune response. CD4+ or CD8+ T cells restrain viral replication and prevent disease, but do not eliminate the latently infected host cell. How do latently infected cells pose a health risk from a potential reactivation? HCMV may be reactivated by immunosuppression, inflammation, differentiation, or critical diseases [9], to begin generating a large number of viral progenies to cause symptoms and diseases, described as the lytic life cycle. When HCMV are reactivated, the transcription of viral genes switches from LAT genes to lytic genes to enhance viral DNA replication and virion production [45]. However, it is not clear how HCMV senses changes in the surrounding immune microenvironment and proliferates. Maintaining the transcription of the LAT gene, maintaining the latent state, inhibiting lysogenic infection, and thus inhibiting the progression of infection, is a good balance for stabilizing the patient.
The immune escape mechanism of HCMV
HCMV can evade an immune response in three ways: evasion from immune recognition receptors, inhibition of immune cell activation, or suppression of effector function (Figure 1).
In terms of evading immune recognition receptors, TLRs comprise a significant signaling pathway required for antiviral defense. The HCMV-encoded glycoproteins US7 and US8 target the TLR3 and TLR4 signaling pathways by promoting the degradation of these TLRs; US7 targets TLR3 and TLR4 by a ubiquitin/proteasome system; and US8 promotes TLR3 and TLR4 destabilization. This results in the overall downregulation of TLR-mediated innate antiviral response [68]. Recognition of HCMV DNA by the cytosolic sensor cyclic GMP/AMP synthase (cGAS) initiates stimulator of interferon genes (STING)-dependent innate antiviral responses. However, the HCMV tegument protein UL82 (also known as pp71) has been identified as a negative regulator of STING-dependent antiviral responses. UL82 inhibits the translocation of STING from the endoplasmic reticulum to perinuclear microsomes by disrupting the STING-iRhom2-TRAPb translocation complex. UL82 also impairs the recruitment of TANK-binding kinase 1 (TBK1) and interferon regulatory factory (IRF3) to the STING complex and impairs STING-mediated signaling [69].
The immune modulation induced by HCMV. HCMV entry into the infected cells through the interaction of the trimeric gH/gL/gO complex (TC) with PDGFRα or the binding of the pentameric gH/gL/UL128-131 complex (PC) with Nrp2, etc. There are multiple strategies for the HCMV to achieve immune escape. For instance, US7 and US8 bind both TLR3 and TLR4, facilitating receptor destabilization by distinct mechanisms; US11 inhibits the assembly of FcRn with β2m resulting in the retention of FcRn in the endoplasmic reticulum, consequently blocking FcRn trafficking to the endosome; UL16, UL142 bind to ligands for NKG2D, the natural killer cell-activating receptor; UL18, US6 interfere with the physical association between MHC class I molecules and TAP; US18, US20 downregulation of B7-H6 leads to evasion from NKp30-mediated killing; US21 protein is a viral-encoded ion channel that regulates intracellular Ca2+ homeostasis and protects cells against apoptosis; UL26 downregulates the expression of antiviral genes; UL82 inhibits STING-mediated signaling; UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112; UL145 hijacks Cullin4 to invoke HLTF; miR-UL22A and miR-US5-2 suppress the secretion of TGF-β; gp68, gP34, gpRL13 and gP95 bind to the Fc segment of IgG on the membrane, resulting in the inability of FcγRⅢA receptor on effector cells to bind to the antibody on the target cells and hinder the communication or cross talk between cellular immunity and humoral immunity.
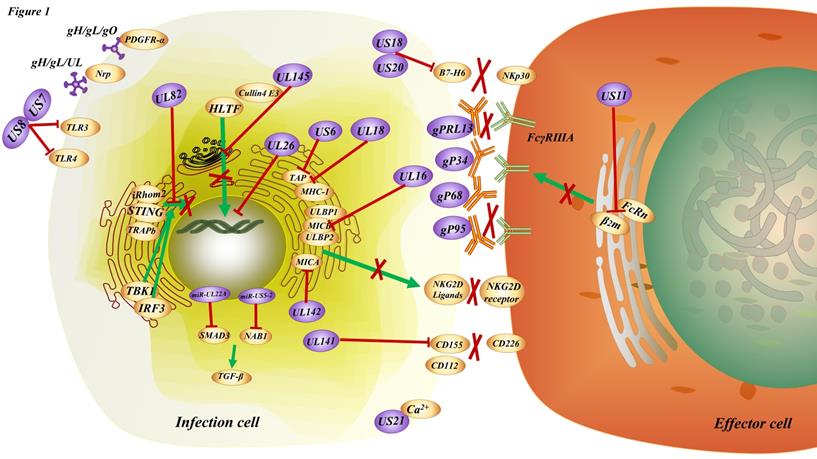
In addition, in order to inhibit immune cell activation, previous studies have shown that there are many HCMV proteins that can inhibit immune cell activation, such as UL16 [70], UL18 [71], US6 [72], US18, US20 [73], US21 [74], UL26 [75], UL140, UL141 [76-78], UL142 [79], US18, US20 [80] even some miRNAs [81, 82]. Recent studies have shown that HCMV infection increases the expression of immune checkpoint genes encoding PD-L1, PD-L2, PD-1, CD80, CD86, Tim-3, LAG3, as well as other T-cell markers such as CD4 and CD8A in tumors along the gastrointestinal tract, including the esophagus, stomach, and intestine [83]. By examining lymphocytes isolated from subjects at the time of viremia, it was found that T cells from individuals with short telomeres proliferated less, produced fewer cytokines, and had less induction of the transcription factor T-box expressed in T cells(T-bet) when stimulated with viral peptides [84]. Helicase-like transcription factor (HLTF), a DNA helicase important for DNA repair, potently inhibits early viral gene expression but is rapidly degraded during infection. The HCMV protein UL145 facilitates HLTF degradation by recruiting the Cullin4 E3 ligase complex and additionally targets tumor protein p53-binding protein 1 (TP53BP1) to invoke this immune evasion strategy in the cytoplasm.
HCMV mainly breaks the link, or the interaction or the communication or the cross talk between cellular immunity and humoral immunity by inhibiting the engagement of HCMV-specific antibody Fc fragments to FcR, and then suppressing effector function. Antibody-dependent cellular phagocytosis (ADCP) occurs upon engagement of virus-specific antibody Fc fragments to FcR, resulting in the cytotoxic killing of infected cells and whole virion degradation. HCMV also encode their own viral FcRs, which recognize the Fc regions of host immunoglobulins. Mimicking host FcRs, vFcRs enable herpesviruses to reduce and evade antiviral immune responses [85]. In addition, gp68 [86], gp34 [87], gpRL13, and gp95 [88, 89], were found to be expressed on the membrane surface of HCMV infected cells, and bind to the Fc segment of IgG on the membrane [85], resulting in the inability of FcγRIIIA receptor on NK cells to bind to the antibody on the target cells and hinder the bridge between cellular immunity and humoral immunity [90, 91]. One study proposed that the HCMV glycoprotein US11 inhibits neonatal Fc receptor functions (FcRn), causing its degradation in the endoplasmic reticulum, which may dampen mucosal and maternal immunity and reduce IgG half-life in blood and tissues, ultimately helping HCMV to escape antibody-mediated immunity [92].
In fact, there are multiple strategies for the “tricky” HCMV to achieve immune escape. At the same time, the elucidation of these immunosuppressive strategies of HCMV also provides ideas for controlling the progression of infection.
Current status of immunotherapy for HCMV
Immunotherapy is the treatment of diseases by artificially enhancing or suppressing the immune function of the body. Immunotherapy has been widely used in the treatment of a large number of cancers, and has achieved good results in different cancers such as the brain [93, 94], breast [95], lung [96], and ovarian cancer [97]. At the same time, it must be admitted that there are still many areas for improvement in immunotherapy. HCMV, which is latent in cells, has been co-evolving with humans over the millenary, and the efficacy of antiviral drug therapy seems suboptimal. With the advancement of immunotherapy, an increasing number of researchers have applied immunotherapy for HCMV infection (Figure 2).
Vaccines: prophylactic and therapeutic
Several types of HCMV vaccine candidates have been developed or are currently under development, including first-generation vaccines (live-attenuated Towne), second-generation vaccines (gB protein-based, peptide vaccines, virus-like particles), and third-generation vaccines (nucleic acid vaccines) [98].
The first-generation vaccine, Towne, attenuated by serial in vitro passaging, seems to have lost its ability for persistent immune induction, as indicated by gradually declining T lymphocyte responses [99]. The Towne vaccine does not cause local or systemic reactions and has an faultless safety record [100, 101]. However, seronegative kidney transplant recipients who received the Towne vaccine were partially protected against HCMV disease, but not against wild-type HCMV infection [102]. The Toledo genome contains mutations disrupting RL13, UL9, UL36, and UL128, and a 14-kb inversion of UL/b' sequences encoding UL130 to UL148 [103]. Thus, due to disruption of UL128, Toledo resembles Towne by not expressing the PC and consequently lacks epithelial or endothelial cell tropism.
The second-generation vaccine, based on early investigation, the chimeras Towne and Toledo of the magnitude and duration of the serologic responses were not notably more rapid or more robust [103]. To date, the most extensively studied vaccine is a subunit vaccine based on the viral envelope glycoprotein B (gpUL55) and tegument phosphoprotein 65(PP65). gB is an essential glycoprotein that plays a crucial role in virus binding to the cell surface, therefore it has been proposed as a target for the development of recombinant vaccines [104]. Furthermore, pp65 is regarded as a significant target of cytotoxic T cell (CTL) [105, 106], and the most likely vaccine target candidate to induce CTL-mediated protection against HCMV diseases [107]. The subunit gB vaccine was developed in the late 1980s and is composed of a Chinese hamster ovary (CHO) cell-derived protein admixed with an oil-in-water emulsion [108]; such the MF59-adjuvanted gB protein subunit vaccine (gB/MF59) is still an important research topic [109, 110]. In the design of the gB/MF59 vaccine, gB was truncated at the transmembrane domain, and the furin protease site was deleted to facilitate purification from CHO cell supernatants. The vaccine was found to be safe and immunogenic in phase I studies [111] and can enhance the neutralizing potency of antibodies against HCMV gB and immune sera under complement enhancement [112]. Acanarypox vector-expressing CMV phosphoprotein 65 is the first recombinant vaccine to elicit CMV-specific CTL responses in humans [113]. Ad-gBCMVpoly is a novel chimeric vaccine based on a replication-deficient adenovirus that encodes a truncated form of CMV-encoded gB antigen and multiple CMV T-cell epitopes from eight different CMV antigens, restricted through multiple human leukocyte antigen (HLA) class I and class II alleles, as a single fusion protein [114]. An enveloped virus-like particle (eVLP) vaccine expressing full-length or chimeric HCMV gB protein was generated by Variation Biotechnologies Incorporated (VBI) laboratories, where the extracellular domain (ECD) of gB is membrane-anchored using the transmembrane and cytoplasmic domains of the vesicular stomatitis virus G protein [115]. eVLP was shown to induce a neutralizing antibody response 10-fold higher than its soluble recombinant protein counterpart [115]. Therefore, a vaccine against the combination of gB, pp65, or US28 (which seems to be a more significant antigen of HCMV) may be more efficacious.
The third-generation vaccine ASP0113 (Astellas Pharma, Tokyo, Japan), a DNA vaccine containing two plasmids encoding CMV antigens (gB aimed at inducing the production of antibodies against CMV and PP65 inducing T-cell mediated responses), was found to be safe and well tolerated in Japanese recipients undergoing allogeneic hematopoietic stem-cell transplantation (HCT). VCL-CB01, a candidate CMV DNA vaccine that contains plasmids encoding CMV pp65 and gB to induce cellular and humoral immune responses, is formulated with poloxamer CRL1005 and benzalkonium chloride to enhance immune responses [116]. The data regarding CMVPepVax, a novel HCMV peptide vaccine formulated with the TLR9 agonist adjuvant PF03512676, provide proof of concept that an HCMV vaccine, in the HCT setting, can increase the number of pp65-specific CD8 T cells, protect from HCMV reactivation, and reduce the use of antivirals [117]. VCL-CT02, a trivalent HCMV DNA vaccine consisting of three plasmids, VCL-6368 (encoding pp65), VCL-6365 (encoding exon 2 and exon 4 of the IE1 gene), and VCL-6520 (encoding the extracellular domain of CMV Gb), appears to safely prime for a memory response to CMV antigens observed after administration of a live, attenuated CMV, and the strength of CMV antigen-specific immune response correlates with the priming effect of the DNA vaccine as measured by a cultured IFN-γ ELISPOT assay [118].
The current prevention and therapy strategies for HCMV infection. With respect to vaccines to prevent HCMV infection, inactivated virus (e.g., Towne, Toledo) weakened by a series of in vitro subcultures and subunit vaccines (e.g., gB/MF59), DNA vaccines (e.g., CMVPepVax, e.g., CMVPepVax, Chimeras, ASP0133, VCL-CB01), have been developed. When these vaccines are injected in the body, they can activate lymphocytes to kill the infected cell antigen-presenting cells (e.g., dendritic cells, macrophages, monocytes). A classic common antiviral inhibitor (cidofovir, ganciclovir, etc.) is a nucleotide analog that mainly impedes the DNA of HCMV synthesis. The target of checkpoint blockade therapy and targeted antibodies in HCMV infection is still under research. HCMV, as an oncolytic virus, usually infects tumor cells and creates an inflammatory microenvironment by causing infected cells to express molecules that target antigenic determinants and recruit immune cells (such as T lymphocytes, macrophages, monocytes, etc.) to cause infected cell apoptosis. ATC involves extraction of PBMC from the body, isolation of target cells (such as T lymphocytes and NK cells) in vitro genetic modification to immunotherapy cells, including HCMV-specific T cells, TCR-T cells, CAR-T cells, and then multiplication, and finally reinfusion into the body.
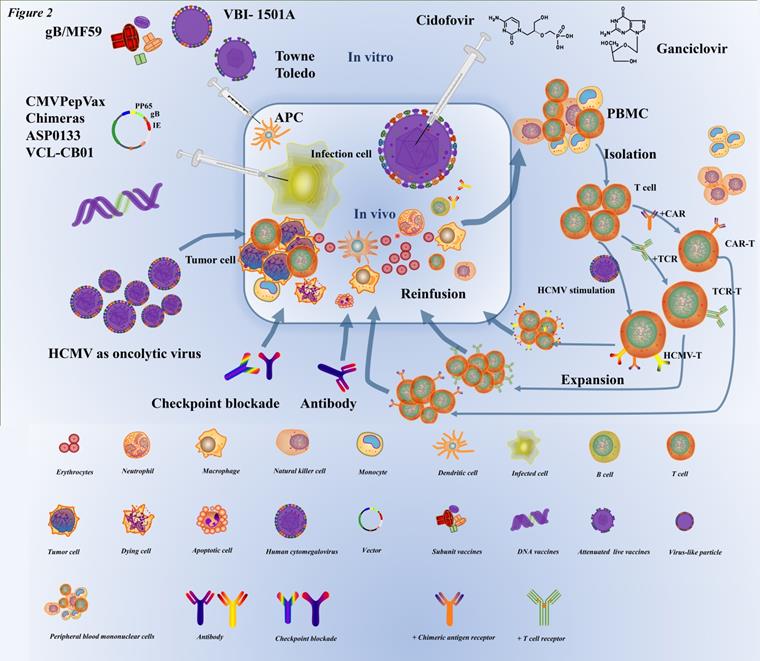
Some scholars propose that an mRNA-based gB vaccine may ultimately prove more efficacious than some second-generation vaccines and increase the durability and breadth of vaccine-elicited antibody responses that can prevent HCMV-associated disease [119]. Nevertheless, the gB vaccine is not entirely reliant on the classic biological activity of neutralization [120]. These potent vaccines are a composite of anti-gB and other major glycoprotein targets, including the trimer gH/gL/gO and the pentameric complex, which may explain the success of the gB HCMV vaccine. However, many molecules, including IE1, IE2, and pp65, have been used as targets for the development of CMV vaccines and the application of vaccine-based immunotherapy [121-123].
Adoptive cell therapy
Adoptive cell therapy (ACT) involves the extraction of immune cells from the body, in vitro genetic modification and amplification, and transfusion back into the patient [124, 125], as a personalized immunotherapy [126]. At present, there are four adoptive cell therapies that have made great progress in international research, including tumor-infiltrating lymphocytes (TILs), T-cell receptor (TCR)-engineered T cells, chimeric antigen receptor (CAR) T cells [127], and natural killer (NK) cell therapy [125]. In 1992, adoptive CMV-specific T-cell therapy against viral infections achieved its first clinical application [128]. Two different strategies have been developed to enrich, isolate, or purify virus-specific T cells [129]. The one is in vitro stimulation and expansion of virus-specific T cells; autologous dendritic cells (DCs) pulsed with viral lysate were used as antigen-presenting cells (APCs) to stimulate CMV-specific T cells in vitro [130], then cells can either be used for in vitro expansion or isolation and direct infusion into the patient. By stimulation and amplification in vitro, a large number of virus-specific T cells can be obtained from a small amount of blood [129]. The second is the direct selection of virus-specific T-cells. For the direct selection of virus-specific T cells, donor white blood cells are isolated ex vivo via peptide-HLA multimers, cytokine-capture method [131, 132], exposure to viral antigens, or methods based on the expression and upregulation of activation molecules on the surface of T cells [133]. Virus-specific T cells are obtained in small amounts and infused into patients where they can expand efficiently and induce viral clearance as well as sustained protection.
ACT has become a therapeutic strategy for HCMV reactivation in patients undergoing allogeneic hematopoietic stem cell transplantation (HSCT) [134, 135] and solid organ transplant (SOT) [136]. For instance, in patients with glioblastoma multiforme (GBM), CMV antigens were found in GBM tissues but not surrounding healthy brain cells [15], suggesting that HCMV plays a key role in glioblastoma and has implications for immunotherapeutic strategies [137]. HCMV promotes murine glioblastoma growth via pericyte recruitment and angiogenesis. A study has shown direct killing of primary GBM cells by autologous HCMV-specific T cells [138], and some investigators developed a novel adoptive immunotherapy approach targeting CMV antigens for patients with recurrent GBM, and experimental results showed that autologous T-cell therapy was completely safe and associated with extended progression-free survival in 4 out of 10 patients [139]. Furthermore, adoptive transfer of virus-specific T cells (VSTs) to achieve antiviral protection for patients treated with allogeneic HSCT resulted in response rates for HCMV of 94% [140]. These results also demonstrate that CMV-specific T cells can effectively target glioblastoma cells for immune killing and support the theoretical basis for the development of CMV directed immunotherapy in patients [138]. Bringing forward strategies of adoptive T-cells promoted the application of transplantation in the treatment of refractory viral infections after HSCT [129]. In clinical application, a 21-year-old female patient with acute myeloid leukemia (AML), was treated with adoptive HCMV-specific T cells from her HLA-haploidentical sister, indicating that HCMV replication could be intermittently controlled by VSTs from an HCMV-positive donor [141]. In recent years, there has been numerous clinical evidence for adoptive HCMV-specific T cell immunotherapy that has achieved satisfactory therapeutic effects [134, 135, 142-144]. These studies strongly suggest that adoptive cellular immunotherapy is a safe and effective approach for treating cancer patients with severe HCMV infections in the future. Indeed, the application of immunotherapy not only provided a new method to anti-CMV strategies, but it is also worthy to be considered in other treatments of refractory viral infections, such as Epstein-Barr virus (EBV), adenovirus (AdV), and even multivirus infections [135, 143, 144].
Oncolytic viruses
CMV is a unique oncolytic virus that is produced by genetic modification of some virulent viruses existing in nature. It uses the inactivation or defect of tumor suppressor genes in target cells to specifically recognize and infect tumor cells, replicate in large quantities, and eventually destroy tumor cells. It also stimulates an immune response that attracts more immune cells to continue killing the remaining cancer cells, and this is a growing field aimed to identify new therapies for treating cancer. Recently, there has been interest in developing vaccines based on CMV, which can induce large numbers of CD8+ T cells that are specific for an epitope that the virus encodes, better known as memory inflation [53, 145-148], thus it can be manipulated to express genes of interest for vaccination [146]. CMV vector expressing the NKG2D ligand RAE-1γ, as a CD8+ T-cell-based anti-malignancy vaccine, could delay tumor growth and even provide complete protection against tumor attack during prevention and treatment [149].
For a lot of “cold tumors,” tumors with low immunogenicity, the response to traditional methods such as radiotherapy, chemotherapy, and immunotherapy is limited [150]; Recent studies have demonstrated that the response is correlated with T cell infiltration and an “inflamed” tumor phenotype [151, 152]. Thus, some scholars have applied CMV-based vaccine methods for converting “non-inflamed” to “inflamed” tumors combined with adoptive T cellar immunotherapy to treat this subset of patients. For instance, MCMVgp100KGP vaccine targeting melanoma gp100 antigen was generated in a laboratory utilizing recombinant murine CMV as a vaccine carrier, gp100 specific CD8+ T cells were activated by the vaccine to effectively protect mice against highly aggressive lung B16-F10 melanoma [153]. Furthermore, MCMVgp100KGP vaccine combined with adoptive T cell therapy have also showed CMV-based vaccines are effective therapies against immunosuppressive solid tumors [154]. The reason why recombinant CMV as a vaccine vector is that CMV-specific CD8+ effector/effector memory T cell populations are large[55], highly dynamic [155], strong cytotoxic effect [156], and have the effect of a longer duration [155, 156]. Therefore, using HCMV as an oncolytic virus to activate the immune system response is beneficial for both the anti-tumor and potentially antiviral response, and ultimately alleviates the disease and may cure the patient.
Whether the CMV can be used as an oncolytic virus is a question that needs to be considered and solved. We have to consider whether CMV has oncogenic properties, infects other normal cells, and produces severe viral infections that exacerbate the disease.
Checkpoint blockade therapy and targeted antibodies
HCMV viremia in renal transplant recipients also appears to upregulate PD-1 expression on CD4 T cells [157]. Studies have indicated that the use of immune checkpoint inhibitors (ICIs) can restore immune function and cause an immune response to CMV antigen when the infection is still latent [158]. The pS-CIFT-aPD-1 is a vector for the expression of the anti-programmed cell death protein 1 (anti-PD-1) antibody gene under the control of a chimeric promoter composed of the CMV enhancer, the core IFN-γ promoter and human T-lymphotropic virus long terminal repeat sequence (TLTR), which co-transfected with the CAR construct into T cells showed increased production of anti-PD-1 antibodies, increased release of IFN-γ, greater T cell activation, and superior antitumor activity [159].
Targeted antibodies are antibodies designed to target antigens on cancer cells, primarily to disrupt the unrestricted growth and proliferation of infected cells. One approach uses antibody peptide epitope conjugates (APECs) to deliver suitable antigens to the tumor surface, which directs pre-existing CMV immunity against tumor cells and activates specifically CMV-reactive effector T cells, whereas a bispecific T-cell engager activates both effector and regulatory T cells [160]. To eliminate the virus, it is more important to activate cellular immunity, whereas antibodies play crucial roles in preventing HCMV spreading in the blood.
Conclusions and future directions
In general, it is obvious that the development of vaccines designed to target HCMV proteins as antigens and stimulate the body to produce effective antibodies and immunocytes has still a long way to go. In adoptive immunotherapy, HCMV is often used as a target antigen to activate anti-CMV-specific T lymphocytes, thereby killing tumor cells infected with CMV. In terms of immune checkpoints, HCMV infection upregulates the expression of genes encoding immune checkpoint genes as well as other immunocyte markers. Although antibody therapy is very limited for CMV because of its intracellular presence in a latent form, interrupting the FcR-associated immune escape pathway and targeting immune checkpoints are promising strategies. In oncolytic virus therapy, CMV is often used as a vector to express oncogenes and induce the anti-tumor ability of T lymphocytes. Of course, due to safety concerns, research in the area of oncolytic viruses is cautious, and the existing research is based on cell and animal models, instead of patients.
HCMV continues to be one of the most significant pathogens affecting the short-term and long-term outcomes of immunocompetent and immunocompromised patients. Firstly, HCMV asymptomatic latent infection and its severity once reactivated are the main obstacles of fundamental research and clinical therapeutics. Considering standardization and personalized immunotherapy, it is difficult to design a relatively perfect immunotherapy regimen to account for the particular complexity of commonly infected patients, different organ transplants, the heterogeneity of tumors (hot and cold tumors), and co-infection with other pathogens simultaneously. Secondly, identification of targets for HCMV immunotherapy, clinically meaningful inclusion criteria, and markers of immune responses, such as peripheral blood cell changes and inflammatory cytokines (IL-6, IFN-γ, TNF-α, IFN-α/β, etc.) is crucially important. Thirdly, the efficacy of single-target immunotherapy or single immunotherapy is limited. However, multi-target or multi-combination immunotherapy may cause unpredictable inflammatory cell infiltration, even cytokine storm, which are worthy of consideration. Finally, advances in immunotherapy technology coupled with the recent fundamental advances in the understanding of HCMV infection have created opportunities for the development of effective immunotherapy for HCMV infection. It is likely that combinatorial regimens with complementary mechanisms of action are required to achieve a broad and durable anti-HCMV benefit.
Abbreviations
HCMV: human cytomegalovirus; HHV-5: human herpesvirus 5; CMV: cytomegalovirus; AIDS: Acquired Immune Deficiency Syndrome; GCV: ganciclovir; CDV: Cidofovir; FOS: Foscavir; dsDNA: double-stranded DNA; UL: unique sequence of long component; US: unique sequence of short component; TRL: terminal repeat of long component; IRL: inverted repeat of long component; IRS: inverted repeat of short component; TRS: terminal repeat of short component; IE: immediate-early gene; E: early gene; L: late gene; gB: Glycoprotein B; gH: glycoprotein H; gL: glycoprotein L; gM: glycoprotein M; gN: glycoprotein N; IL-1β: interleukin-1β; TC: trimeric gH/gL/gO complex; PDGFRα: platelet-derived growth factor receptor alpha; PC: pentameric gH/gL/UL128-131 complex; Nrp2: neuropilin-2; MTs: microtubules; MIEP: major immediate early promoter; EMT: epithelial mesenchymal transition; ROCK: Rho-associated coiled-coil kinase; LAT: latency-associated transcript; LUNA: latent undefined nuclear antigen; ERK: extracellular signal-regulated kinase; MAPK: mitogen activated protein kinase; TGF-β: transforming growth factor-β; BAFF-R: B cell-activating factor; NK cells: natural killer cells; TLR: toll-like receptors; STING: stimulator of interferon genes; cGAS: cyclic GMP/AMP synthase; TBK1: TANK-binding kinase 1; IRF3: interferon regulatory factory; PD-1: programmed cell death protein 1; PD-L1: programmed death ligand-1; TIM-3: T cell immunoglobulin domain and mucin domain-3; LAG-3: Lymphocyte-activation-gene-3; HLTF: Helicase-like transcription factor; TP53BP1: tumor protein p53-binding protein 1; ADCP: antibody-dependent cellular phagocytosis; FcRn: Fc receptor functions; PP65: phosphoprotein 65; CTL: cytotoxic T cell; CHO cell: Chinese hamster ovary; gB/MF59: MF59-adjuvanted gB protein subunit vaccine; HLA: human leukocyte antigen; eVLP: enveloped virus-like particle; HCT: hematopoietic stem-cell transplantation; ACT: adoptive cell therapy; TILs: tumor-infiltrating lymphocytes; TCR: T-cell receptor; CAR: chimeric antigen receptor; DC: dendritic cells; APCs: antigen-presenting cells; HSCT: hematopoietic stem cell transplantation; SOT: solid organ transplant; GBM: glioblastoma multiforme; VSTs: virus-specific T cells; EBV: Epstein-Barr virus; AdV: adenovirus; TLTR: T-lymphotropic virus long terminal repeat sequence; APECs: antibody peptide epitope conjugates; TAP: transporter associated with antigen processing.
Acknowledgements
We thank the support from the National Natural Science Foundation of China (82073096), Key Research and Development Plan of Hunan Province (2020SK2053).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther. 2003;98:269-97
2. Craig JM, Macauley JC, Weller TH, Wirth P. Isolation of intranuclear inclusion producing agents from infants with illnesses resembling cytomegalic inclusion disease. Proc Soc Exp Biol Med. 1957;94:4-12
3. Zuhair M, Smit GSA, Wallis G, Jabbar F, Smith C, Devleesschauwer B. et al. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev Med Virol. 2019;29:e2034
4. Kawai K, Itoh H. Congenital Cytomegalovirus Infection. N Engl J Med. 2018;379:e21
5. Teira P, Battiwalla M, Ramanathan M, Barrett AJ, Ahn KW, Chen M. et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: a CIBMTR analysis. Blood. 2016;127:2427-38
6. Schmidt-Hieber M, Tridello G, Ljungman P, Mikulska M, Knelange N, Blaise D. et al. The prognostic impact of the cytomegalovirus serostatus in patients with chronic hematological malignancies after allogeneic hematopoietic stem cell transplantation: a report from the Infectious Diseases Working Party of EBMT. Ann Hematol. 2019;98:1755-63
7. Ziemann M, Thiele T. Transfusion-transmitted CMV infection - current knowledge and future perspectives. Transfus Med. 2017;27:238-48
8. Weisberg SP, Staley EM, Williams LA 3rd, Pham HP, Bachegowda LS, Cheng YH. et al. Survey on Transfusion-Transmitted Cytomegalovirus and Cytomegalovirus Disease Mitigation. Arch Pathol Lab Med. 2017;141:1705-11
9. Dupont L, Reeves MB. Cytomegalovirus latency and reactivation: recent insights into an age old problem. Rev Med Virol. 2016;26:75-89
10. Korver AM, Smith RJ, Van Camp G, Schleiss MR, Bitner-Glindzicz MA, Lustig LR. et al. Congenital hearing loss. Nat Rev Dis Primers. 2017;3:16094
11. Purswani MU, Russell JS, Dietrich M, Malee K, Spector SA, Williams PL. et al. Birth Prevalence of Congenital Cytomegalovirus Infection in HIV-Exposed Uninfected Children in the Era of Combination Antiretroviral Therapy. J Pediatr. 2020;216:82-7 e2
12. Gronborg HL, Jespersen S, Honge BL, Jensen-Fangel S, Wejse C. Review of cytomegalovirus coinfection in HIV-infected individuals in Africa. Rev Med Virol. 2017;27:e1907
13. Marty FM, Ljungman P, Chemaly RF, Maertens J, Dadwal SS, Duarte RF. et al. Letermovir Prophylaxis for Cytomegalovirus in Hematopoietic-Cell Transplantation. N Engl J Med. 2017;377:2433-44
14. Dioverti MV, Razonable RR. Cytomegalovirus. Microbiol Spectr. 2016;4:4.20
15. Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH. et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347-50
16. Yang Z, Tang X, Meng G, Benesch MGK, Mackova M, Belon AP. et al. Latent Cytomegalovirus Infection in Female Mice Increases Breast Cancer Metastasis. Cancers (Basel). 2019;11:447
17. Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561-4
18. Hostetler KY. Synthesis and early development of hexadecyloxypropylcidofovir: an oral antipoxvirus nucleoside phosphonate. Viruses. 2010;2:2213-25
19. Sahu PK, Umme T, Yu J, Nayak A, Kim G, Noh M. et al. Selenoacyclovir and Selenoganciclovir: Discovery of a New Template for Antiviral Agents. J Med Chem. 2015;58:8734-8
20. Sun K, Xu H, Hilfinger JL, Lee KD, Provoda CJ, Sabit H. et al. Improved Protease-Targeting and Biopharmaceutical Properties of Novel Prodrugs of Ganciclovir. Mol Pharm. 2018;15:410-9
21. Chen SJ, Wang SC, Chen YC. Antiviral Agents as Therapeutic Strategies Against Cytomegalovirus Infections. Viruses. 2019;12(1):21
22. Gibson W. Structure and formation of the cytomegalovirus virion. Curr Top Microbiol Immunol. 2008;325:187-204
23. Griffiths PD, Grundy JE. Molecular biology and immunology of cytomegalovirus. Biochem J. 1987;241:313-24
24. Balazs Z, Tombacz D, Szucs A, Csabai Z, Megyeri K, Petrov AN. et al. Long-Read Sequencing of Human Cytomegalovirus Transcriptome Reveals RNA Isoforms Carrying Distinct Coding Potentials. Sci Rep. 2017;7:15989
25. Britt WJ, Prichard MN. New therapies for human cytomegalovirus infections. Antiviral Res. 2018;159:153-74
26. Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Chee MS. et al. The DNA sequence of the human cytomegalovirus genome. DNA Seq. 1991;2:1-12
27. Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA. et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A. 2003;100:14976-81
28. Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L. et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78:10960-6
29. Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol. 2000;74:7720-9
30. Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V. et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003;100:14223-8
31. Vanarsdall AL, Johnson DC. Human cytomegalovirus entry into cells. Curr Opin Virol. 2012;2:37-42
32. Schleiss MR. Recombinant cytomegalovirus glycoprotein B vaccine: Rethinking the immunological basis of protection. Proc Natl Acad Sci U S A. 2018;115:6110-2
33. Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A. 2005;102:18153-8
34. Straschewski S, Patrone M, Walther P, Gallina A, Mertens T, Frascaroli G. Protein pUL128 of human cytomegalovirus is necessary for monocyte infection and blocking of migration. J Virol. 2011;85:5150-8
35. Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol. 2006;87:2451-60
36. Nguyen CC, Kamil JP. Pathogen at the Gates: Human Cytomegalovirus Entry and Cell Tropism. Viruses. 2018;10:704
37. Wu Y, Prager A, Boos S, Resch M, Brizic I, Mach M. et al. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-alpha as a key for entry. PLoS Pathog. 2017;13:e1006281
38. Martinez-Martin N, Marcandalli J, Huang CS, Arthur CP, Perotti M, Foglierini M. et al. An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell. 2018;174:1158-71 e19
39. Sinzger C, Kahl M, Laib K, Klingel K, Rieger P, Plachter B. et al. Tropism of human cytomegalovirus for endothelial cells is determined by a post-entry step dependent on efficient translocation to the nucleus. J Gen Virol. 2000;81:3021-35
40. Ogawa-Goto K, Tanaka K, Gibson W, Moriishi E, Miura Y, Kurata T. et al. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J Virol. 2003;77:8541-7
41. Kostopoulou ON, Wilhelmi V, Raiss S, Ananthaseshan S, Lindstrom MS, Bartek J. et al. Human cytomegalovirus and Herpes Simplex type I virus can engage RNA polymerase I for transcription of immediate early genes. Oncotarget. 2017;8:96536-52
42. Eliyahu E, Tirosh O, Dobesova M, Nachshon A, Schwartz M, Stern-Ginossar N. Rho-Associated Coiled-Coil Kinase 1 Translocates to the Nucleus and Inhibits Human Cytomegalovirus Propagation. J Virol. 2019;93:e00453-19
43. Popescu I, Mannem H, Winters SA, Hoji A, Silveira F, McNally E. et al. Impaired Cytomegalovirus Immunity in Idiopathic Pulmonary Fibrosis Lung Transplant Recipients with Short Telomeres. Am J Respir Crit Care Med. 2019;199:362-76
44. Moussawi FA, Kumar A, Pasquereau S, Tripathy MK, Karam W, Diab-Assaf M. et al. The transcriptome of human mammary epithelial cells infected with the HCMV-DB strain displays oncogenic traits. Sci Rep. 2018;8:12574
45. Chen Y-C, Sheng J, Trang P, Liu F. Potential Application of the CRISPR/Cas9 System against Herpesvirus Infections. Viruses. 2018;10(6):291
46. Lee SH, Albright ER, Lee JH, Jacobs D, Kalejta RF. Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein. Sci Adv. 2015;1:e1501164
47. Reeves M, Woodhall D, Compton T, Sinclair J. Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J Virol. 2010;84(14):7185-94
48. Bego M, Maciejewski J, Khaiboullina S, Pari G, St Jeor S. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J Virol. 2005;79:11022-34
49. Krishna BA, Poole EL, Jackson SE, Smit MJ, Wills MR, Sinclair JH. Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection. mBio. 2017;8(6):e01754-17
50. Krishna BA, Miller WE, O'Connor CM. US28: HCMV's Swiss Army Knife. Viruses. 2018;10(8):445
51. Zischke J, Mamareli P, Pokoyski C, Gabaev I, Buyny S, Jacobs R. et al. The human cytomegalovirus glycoprotein pUL11 acts via CD45 to induce T cell IL-10 secretion. PLoS Pathog. 2017;13:e1006454
52. Mason GM, Poole E, Sissons JG, Wills MR, Sinclair JH. Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc Natl Acad Sci U S A. 2012;109:14538-43
53. Söderberg-Nauclér C, Fish KN, Nelson JA. Reactivation of Latent Human Cytomegalovirus by Allogeneic Stimulation of Blood Cells from Healthy Donors. Cell. 1997;91:119-26
54. Kew VG, Yuan J, Meier J, Reeves MB. Mitogen and stress activated kinases act co-operatively with CREB during the induction of human cytomegalovirus immediate-early gene expression from latency. PLoS Pathog. 2014;10:e1004195
55. Klenerman P, Oxenius A. T cell responses to cytomegalovirus. Nat Rev Immunol. 2016;16:367-77
56. Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. Human NK cells can control CMV infection in the absence of T cells. Blood. 2008;112:914-5
57. Cooley S, Parham P, Miller JS. Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood. 2018;131:1053-62
58. Nikitina E, Larionova I, Choinzonov E, Kzhyshkowska J. Monocytes and Macrophages as Viral Targets and Reservoirs. Int J Mol Sci. 2018;19:2821
59. Farrell HE, Bruce K, Ma J, Davis-Poynter N, Stevenson PG. Human cytomegalovirus US28 allows dendritic cell exit from lymph nodes. J Gen Virol. 2018;99:1509-14
60. Liu J, Jardetzky TS, Chin AL, Johnson DC, Vanarsdall AL. The Human Cytomegalovirus Trimer and Pentamer Promote Sequential Steps in Entry into Epithelial and Endothelial Cells at Cell Surfaces and Endosomes. J Virol. 2018;92(21):e01336-18
61. Dennis EA, Smythies LE, Grabski R, Li M, Ballestas ME, Shimamura M. et al. Cytomegalovirus promotes intestinal macrophage-mediated mucosal inflammation through induction of Smad7. Mucosal Immunol. 2018;11:1694-704
62. Hammer Q, Romagnani C. About Training and Memory: NK-Cell Adaptation to Viral Infections. Adv Immunol. 2017;133:171-207
63. Min CK, Shakya AK, Lee BJ, Streblow DN, Caposio P, Yurochko AD. The Differentiation of Human Cytomegalovirus Infected-Monocytes Is Required for Viral Replication. Front Cell Infect Microbiol. 2020;10:368
64. Xu H, Dong P, Ma X, Song D, Xue D, Xu R. et al. B cell-activating factor regulates the survival of B lymphocytes infected with human cytomegalovirus. Immunol Lett. 2017;187:1-6
65. HoHsieh A, Wang CM, Wu YJ, Chen A, Chang MI, Chen JY. B cell epitope of human cytomegalovirus phosphoprotein 65 (HCMV pp65) induced anti-dsDNA antibody in BALB/c mice. Arthritis Res Ther. 2017;19:65
66. Biolatti M, Dell'Oste V, Scutera S, Gugliesi F, Griffante G, De Andrea M. et al. The Viral Tegument Protein pp65 Impairs Transcriptional Upregulation of IL-1beta by Human Cytomegalovirus through Inhibition of NF-kB Activity. Viruses. 2018;10:567
67. Link EK, Brandmuller C, Suezer Y, Ameres S, Volz A, Moosmann A. et al. A synthetic human cytomegalovirus pp65-IE1 fusion antigen efficiently induces and expands virus specific T cells. Vaccine. 2017;35:5131-9
68. Park A, Ra EA, Lee TA, Choi HJ, Lee E, Kang S. et al. HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways. Nat Commun. 2019;10:4670
69. Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM. et al. Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe. 2017;21:231-43
70. Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC. et al. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427-39
71. Farrell HE, Vally H, Lynch DM, Fleming P, Shellam GR, Scalzo AA. et al. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 1997;386:510-4
72. Kim Y, Park B, Cho S, Shin J, Cho K, Jun Y. et al. Human cytomegalovirus UL18 utilizes US6 for evading the NK and T-cell responses. PLoS Pathog. 2008;4:e1000123
73. Charpak-Amikam Y, Kubsch T, Seidel E, Oiknine-Djian E, Cavaletto N, Yamin R. et al. Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7-H6 by the viral genes US18 and US20. Sci Rep. 2017;7:8661
74. Luganini A, Di Nardo G, Munaron L, Gilardi G, Fiorio Pla A, Gribaudo G. Human cytomegalovirus US21 protein is a viroporin that modulates calcium homeostasis and protects cells against apoptosis. Proc Natl Acad Sci U S A. 2018;115:E12370-E7
75. Goodwin CM, Schafer X, Munger J. UL26 Attenuates IKKbeta-Mediated Induction of Interferon-Stimulated Gene (ISG) Expression and Enhanced Protein ISGylation during Human Cytomegalovirus Infection. J Virol. 2019;93:e01052-19
76. Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C. et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6:181-8
77. Prod'homme V, Sugrue DM, Stanton RJ, Nomoto A, Davies J, Rickards CR. et al. Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J Gen Virol. 2010;91:2034-9
78. Dufour F, Rattier T, Shirley S, Picarda G, Constantinescu AA, Morle A. et al. N-glycosylation of mouse TRAIL-R and human TRAIL-R1 enhances TRAIL-induced death. Cell Death Differ. 2017;24:500-10
79. Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun. 2006;346:175-81
80. Fielding CA, Weekes MP, Nobre LV, Ruckova E, Wilkie GS, Paulo JA. et al. Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. Elife. 2017 6
81. Moller R, Schwarz TM, Noriega VM, Panis M, Sachs D, Tortorella D. et al. miRNA-mediated targeting of human cytomegalovirus reveals biological host and viral targets of IE2. Proc Natl Acad Sci U S A. 2018;115:1069-74
82. Hancock MH, Crawford LB, Pham AH, Mitchell J, Struthers HM, Yurochko AD. et al. Human Cytomegalovirus miRNAs Regulate TGF-beta to Mediate Myelosuppression while Maintaining Viral Latency in CD34(+) Hematopoietic Progenitor Cells. Cell Host Microbe. 2020;27:104-14 e4
83. Cao S, Wylie KM, Wyczalkowski MA, Karpova A, Ley J, Sun S. et al. Dynamic host immune response in virus-associated cancers. Commun Biol. 2019;2:109
84. Greenland JR. Where the Chromosome Ends: Telomeres and Cytomegalovirus Risk in Lung Transplant Recipients. Am J Respir Crit Care Med. 2019;199:265-7
85. Jenks JA, Goodwin ML, Permar SR. The Roles of Host and Viral Antibody Fc Receptors in Herpes Simplex Virus (HSV) and Human Cytomegalovirus (HCMV) Infections and Immunity. Front Immunol. 2019;10:2110
86. Ndjamen B, Joshi DS, Fraser SE, Bjorkman PJ. Characterization of Antibody Bipolar Bridging Mediated by the Human Cytomegalovirus Fc Receptor gp68. J Virol. 2016;90:3262-7
87. Corrales-Aguilar E, Trilling M, Hunold K, Fiedler M, Le VT, Reinhard H. et al. Human cytomegalovirus Fcgamma binding proteins gp34 and gp68 antagonize Fcgamma receptors I, II and III. PLoS Pathog. 2014;10:e1004131
88. Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M. et al. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J Virol. 2002;76:8596-608
89. Cortese M, Calo S, D'Aurizio R, Lilja A, Pacchiani N, Merola M. Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One. 2012;7:e50166
90. Hogarth PM, Pietersz GA. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov. 2012;11:311-31
91. Lilley BN, Ploegh HL, Tirabassi RS. Human cytomegalovirus open reading frame TRL11/IRL11 encodes an immunoglobulin G Fc-binding protein. J Virol. 2001;75:11218-21
92. Liu X, Palaniyandi S, Zhu I, Tang J, Li W, Wu X. et al. Human cytomegalovirus evades antibody-mediated immunity through endoplasmic reticulum-associated degradation of the FcRn receptor. Nat Commun. 2019;10:3020
93. Sampson JH, Gunn MD, Fecci PE, Ashley DM. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20:12-25
94. Sampson JH, Maus MV, June CH. Immunotherapy for Brain Tumors. J Clin Oncol. 2017;35:2450-6
95. Emens LA. Breast Cancer Immunotherapy: Facts and Hopes. Clin Cancer Res. 2018;24:511-20
96. Steven A, Fisher SA, Robinson BW. Immunotherapy for lung cancer. Respirology. 2016;21:821-33
97. Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. 2017;28:viii1-viii7
98. Inoue N, Abe M, Kobayashi R, Yamada S. Vaccine Development for Cytomegalovirus. Adv Exp Med Biol. 2018;1045:271-96
99. Jacobson MA, Sinclair E, Bredt B, Agrillo L, Black D, Epling CL. et al. Antigen-specific T cell responses induced by Towne cytomegalovirus (CMV) vaccine in CMV-seronegative vaccine recipients. J Clin Virol. 2006;35:332-7
100. Jacobson MA, Sinclair E, Bredt B, Agrillo L, Black D, Epling CL. et al. Safety and immunogenicity of Towne cytomegalovirus vaccine with or without adjuvant recombinant interleukin-12. Vaccine. 2006;24:5311-9
101. Pass RF, Burke RL. Development of cytomegalovirus vaccines: prospects for prevention of congenital CMV infection. Semin Pediatr Infect Dis. 2002;13:196-204
102. Plotkin SA, Starr SE, Friedman HM, Brayman K, Harris S, Jackson S. et al. Effect of Towne live virus vaccine on cytomegalovirus disease after renal transplant. A controlled trial. Ann Intern Med. 1991;114:525-31
103. Adler SP, Manganello AM, Lee R, McVoy MA, Nixon DE, Plotkin S. et al. A Phase 1 Study of 4 Live, Recombinant Human Cytomegalovirus Towne/Toledo Chimera Vaccines in Cytomegalovirus-Seronegative Men. J Infect Dis. 2016;214:1341-8
104. Gyulai Z, Endresz V, Burian K, Pincus S, Toldy J, Cox WI. et al. Cytotoxic T lymphocyte (CTL) responses to human cytomegalovirus pp65, IE1-Exon4, gB, pp150, and pp28 in healthy individuals: reevaluation of prevalence of IE1-specific CTLs. J Infect Dis. 2000;181:1537-46
105. Kondo E, Akatsuka Y, Kuzushima K, Tsujimura K, Asakura S, Tajima K. et al. Identification of novel CTL epitopes of CMV-pp65 presented by a variety of HLA alleles. Blood. 2004;103:630-8
106. Akiyama Y, Tai S, Komiyama M, Takikawa M, Ohshita C, Yamamoto A. et al. Characterization of cytomegalovirus pp65-HLA-A24 peptide-specific CTL lines from metastatic melanoma patients. Oncol Rep. 2009;22:185-91
107. Berencsi K, Gyulai Z, Gönczöl E, Pincus S, Cox WI, Michelson S. et al. A canarypox vector-expressing cytomegalovirus (CMV) phosphoprotein 65 induces long-lasting cytotoxic T cell responses in human CMV-seronegative subjects. J Infect Dis. 2001;183:1171-9
108. Pass RF, Duliege AM, Boppana S, Sekulovich R, Percell S, Britt W. et al. A subunit cytomegalovirus vaccine based on recombinant envelope glycoprotein B and a new adjuvant. J Infect Dis. 1999;180:970-5
109. Jenks JA, Nelson CS, Roark HK, Goodwin ML, Pass RF, Bernstein DI. et al. Antibody binding to native cytomegalovirus glycoprotein B predicts efficacy of the gB/MF59 vaccine in humans. Sci Transl Med. 2020;12:eabb3611
110. Baraniak I, Gomes AC, Sodi I, Langstone T, Rothwell E, Atkinson C. et al. Seronegative patients vaccinated with cytomegalovirus gB-MF59 vaccine have evidence of neutralising antibody responses against gB early post-transplantation. EBioMedicine. 2019;50:45-54
111. Frey SE, Harrison C, Pass RF, Yang E, Boken D, Sekulovich RE. et al. Effects of antigen dose and immunization regimens on antibody responses to a cytomegalovirus glycoprotein B subunit vaccine. J Infect Dis. 1999;180:1700-3
112. Li F, Freed DC, Tang A, Rustandi RR, Troutman MC, Espeseth AS. et al. Complement enhances in vitro neutralizing potency of antibodies to human cytomegalovirus glycoprotein B (gB) and immune sera induced by gB/MF59 vaccination. NPJ Vaccines. 2017;2:36
113. Berencsi K, Gyulai Z, Gonczol E, Pincus S, Cox WI, Michelson S. et al. A canarypox vector-expressing cytomegalovirus (CMV) phosphoprotein 65 induces long-lasting cytotoxic T cell responses in human CMV-seronegative subjects. J Infect Dis. 2001;183:1171-9
114. Rist M, Cooper L, Elkington R, Walker S, Fazou C, Tellam J. et al. Ex vivo expansion of human cytomegalovirus-specific cytotoxic T cells by recombinant polyepitope: implications for HCMV immunotherapy. Eur J Immunol. 2005;35:996-1007
115. Kirchmeier M, Fluckiger AC, Soare C, Bozic J, Ontsouka B, Ahmed T. et al. Enveloped virus-like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity. Clin Vaccine Immunol. 2014;21:174-80
116. Wloch MK, Smith LR, Boutsaboualoy S, Reyes L, Han C, Kehler J. et al. Safety and immunogenicity of a bivalent cytomegalovirus DNA vaccine in healthy adult subjects. J Infect Dis. 2008;197:1634-42
117. Nakamura R, Rosa CL, Longmate J, Drake J, Slape C, Zhou Q. et al. Viraemia, immunogenicity, and survival outcomes of cytomegalovirus chimeric epitope vaccine supplemented with PF03512676 (CMVPepVax) in allogeneic haemopoietic stem-cell transplantation: randomised phase 1b trial. Lancet Haematol. 2016;3:e87-e98
118. Jacobson MA, Adler SP, Sinclair E, Black D, Smith A, Chu A. et al. A CMV DNA vaccine primes for memory immune responses to live-attenuated CMV (Towne strain). Vaccine. 2009;27:1540-8
119. Nelson CS, Jenks JA, Pardi N, Goodwin M, Roark H, Edwards W. et al. Human Cytomegalovirus Glycoprotein B Nucleoside-Modified mRNA Vaccine Elicits Antibody Responses with Greater Durability and Breadth than MF59-Adjuvanted gB Protein Immunization. J Virol. 2020;94:e00186-20
120. Baraniak I, Kropff B, Ambrose L, McIntosh M, McLean GR, Pichon S. et al. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A. 2018;115:6273-8
121. Mori T, Kanda Y, Takenaka K, Okamoto S, Kato J, Kanda J. et al. Safety of ASP0113, a cytomegalovirus DNA vaccine, in recipients undergoing allogeneic hematopoietic cell transplantation: an open-label phase 2 trial. Int J Hematol. 2017;105:206-12
122. Kabanova A, Perez L, Lilleri D, Marcandalli J, Agatic G, Becattini S. et al. Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc Natl Acad Sci U S A. 2014;111:17965-70
123. La Rosa C, Longmate J, Martinez J, Zhou Q, Kaltcheva TI, Tsai W. et al. MVA vaccine encoding CMV antigens safely induces durable expansion of CMV-specific T cells in healthy adults. Blood. 2017;129:114-25
124. Guedan S, Ruella M, June CH. Emerging Cellular Therapies for Cancer. Annu Rev Immunol. 2019;37:145-71
125. Lee DA. Cellular therapy: Adoptive immunotherapy with expanded natural killer cells. Immunol Rev. 2019;290:85-99
126. Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62-8
127. Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 2017;168:724-40
128. Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238-41
129. Kaeuferle T, Krauss R, Blaeschke F, Willier S, Feuchtinger T. Strategies of adoptive T -cell transfer to treat refractory viral infections post allogeneic stem cell transplantation. J Hematol Oncol. 2019;12:13
130. Peggs K, Verfuerth S, Mackinnon S. Induction of cytomegalovirus (CMV)-specific T-cell responses using dendritic cells pulsed with CMV antigen: a novel culture system free of live CMV virions. Blood. 2001;97:994-1000
131. Feuchtinger T, Opherk K, Bethge WA, Topp MS, Schuster FR, Weissinger EM. et al. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood. 2010;116:4360-7
132. Feucht J, Opherk K, Lang P, Kayser S, Hartl L, Bethge W. et al. Adoptive T-cell therapy with hexon-specific Th1 cells as a treatment of refractory adenovirus infection after HSCT. Blood. 2015;125:1986-94
133. Khanna N, Stuehler C, Conrad B, Lurati S, Krappmann S, Einsele H. et al. Generation of a multipathogen-specific T-cell product for adoptive immunotherapy based on activation-dependent expression of CD154. Blood. 2011;118:1121-31
134. Neuenhahn M, Albrecht J, Odendahl M, Schlott F, Dossinger G, Schiemann M. et al. Transfer of minimally manipulated CMV-specific T cells from stem cell or third-party donors to treat CMV infection after allo-HSCT. Leukemia. 2017;31:2161-71
135. Withers B, Blyth E, Clancy LE, Yong A, Fraser C, Burgess J. et al. Long-term control of recurrent or refractory viral infections after allogeneic HSCT with third-party virus-specific T cells. Blood Adv. 2017;1:2193-205
136. Beam E, Dioverti V, Razonable RR. Emerging cytomegalovirus management strategies after solid organ transplantation: challenges and opportunities. Curr Infect Dis Rep. 2014;16:419
137. Rahman M, Dastmalchi F, Karachi A, Mitchell D. The role of CMV in glioblastoma and implications for immunotherapeutic strategies. Oncoimmunology. 2019;8:e1514921
138. Nair SK, De Leon G, Boczkowski D, Schmittling R, Xie W, Staats J. et al. Recognition and killing of autologous, primary glioblastoma tumor cells by human cytomegalovirus pp65-specific cytotoxic T cells. Clin Cancer Res. 2014;20:2684-94
139. Schuessler A, Walker DG, Khanna R. Cellular immunotherapy directed against human cytomegalovirus as a novel approach for glioblastoma treatment. Oncoimmunology. 2014;3:e29381
140. Tzannou I, Papadopoulou A, Naik S, Leung K, Martinez CA, Ramos CA. et al. Off-the-Shelf Virus-Specific T Cells to Treat BK Virus, Human Herpesvirus 6, Cytomegalovirus, Epstein-Barr Virus, and Adenovirus Infections After Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol. 2017;35:3547-57
141. Lindemann M, Eiz-Vesper B, Steckel NK, Tischer S, Fiedler M, Heinold A. et al. Adoptive transfer of cellular immunity against cytomegalovirus by virus-specific lymphocytes from a third-party family donor. Bone Marrow Transplant. 2018;53:1351-5
142. Pei XY, Zhao XY, Chang YJ, Liu J, Xu LP, Wang Y. et al. Cytomegalovirus-Specific T-Cell Transfer for Refractory Cytomegalovirus Infection After Haploidentical Stem Cell Transplantation: The Quantitative and Qualitative Immune Recovery for Cytomegalovirus. J Infect Dis. 2017;216:945-56
143. Withers B, Clancy L, Burgess J, Simms R, Brown R, Micklethwaite K. et al. Establishment and Operation of a Third-Party Virus-Specific T Cell Bank within an Allogeneic Stem Cell Transplant Program. Biol Blood Marrow Transplant. 2018;24:2433-42
144. Kallay K, Kassa C, Reti M, Karaszi E, Sinko J, Goda V. et al. Early Experience With CliniMACS Prodigy CCS (IFN-gamma) System in Selection of Virus-specific T Cells From Third-party Donors for Pediatric Patients With Severe Viral Infections After Hematopoietic Stem Cell Transplantation. J Immunother. 2018;41:158-63
145. Holtappels R, Grzimek NK, Simon CO, Thomas D, Dreis D, Reddehase MJ. Processing and presentation of murine cytomegalovirus pORFm164-derived peptide in fibroblasts in the face of all viral immunosubversive early gene functions. J Virol. 2002;76:6044-53
146. Karrer U, Wagner M, Sierro S, Oxenius A, Hengel H, Dumrese T. et al. Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J Virol. 2004;78:2255-64
147. Munks MW, Gold MC, Zajac AL, Doom CM, Morello CS, Spector DH. et al. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J Immunol. 2006;176:3760-6
148. Holtappels R, Pahl-Seibert MF, Thomas D, Reddehase MJ. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J Virol. 2000;74:11495-503
149. Trsan T, Vukovic K, Filipovic P, Brizic AL, Lemmermann NAW, Schober K. et al. Cytomegalovirus vector expressing RAE-1gamma induces enhanced anti-tumor capacity of murine CD8(+) T cells. Eur J Immunol. 2017;47:1354-67
150. Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T. et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462-9
151. Gajewski TF. The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin Oncol. 2015;42:663-71
152. Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T. et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity. 2018;49:178-93 e7
153. Qiu Z, Huang H, Grenier JM, Perez OA, Smilowitz HM, Adler B. et al. Cytomegalovirus-Based Vaccine Expressing a Modified Tumor Antigen Induces Potent Tumor-Specific CD8(+) T-cell Response and Protects Mice from Melanoma. Cancer Immunol Res. 2015;3:536-46
154. Grenier JM, Yeung ST, Qiu Z, Jellison ER, Khanna KM. Combining Adoptive Cell Therapy with Cytomegalovirus-Based Vaccine Is Protective against Solid Skin Tumors. Front Immunol. 2017;8:1993
155. Snyder CM, Cho KS, Bonnett EL, van Dommelen S, Shellam GR, Hill AB. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity. 2008;29:650-9
156. Snyder CM, Cho KS, Bonnett EL, Allan JE, Hill AB. Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS Pathog. 2011;7:e1002295
157. Dirks J, Tas H, Schmidt T, Kirsch S, Gartner BC, Sester U. et al. PD-1 analysis on CD28(-) CD27(-) CD4 T cells allows stimulation-independent assessment of CMV viremic episodes in transplant recipients. Am J Transplant. 2013;13:3132-41
158. Lin X, Lu T, Li S, Xie X, Chen X, Jiang J. et al. Cytomegalovirus infection as an underestimated trigger for checkpoint inhibitor-related pneumonitis in lung cancer: a retrospective study. Clin Transl Oncol. 2020;23(2):389-96
159. Fang Y, Zhang Y, Guo C, Chen C, Gao H, Zhou X. et al. Safety and Efficacy of an Immune Cell-Specific Chimeric Promoter in Regulating Anti-PD-1 Antibody Expression in CAR T Cells. Mol Ther Methods Clin Dev. 2020;19:14-23
160. Millar DG, Ramjiawan RR, Kawaguchi K, Gupta N, Chen J, Zhang S. et al. Antibody-mediated delivery of viral epitopes to tumors harnesses CMV-specific T cells for cancer therapy. Nat Biotechnol. 2020;38:420-5
Author contact
![]() Corresponding author: Minghua Wu, Cancer Research Institute, Central South University, Changsha, Hunan, China. 410078. Tel: 86-731-82355401. E-mail: wuminghua554com; Zuping Zhang, Department of Pathogeny Biology, School of Basic Medical Science, Central South University, Changsha, Hunan, China. 410078. Tel: 86-731-82650556. E-mail: zhangzupingedu.cn.
Corresponding author: Minghua Wu, Cancer Research Institute, Central South University, Changsha, Hunan, China. 410078. Tel: 86-731-82355401. E-mail: wuminghua554com; Zuping Zhang, Department of Pathogeny Biology, School of Basic Medical Science, Central South University, Changsha, Hunan, China. 410078. Tel: 86-731-82650556. E-mail: zhangzupingedu.cn.

 Global reach, higher impact
Global reach, higher impact