Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(15):4493-4513. doi:10.7150/ijbs.66181 This issue Cite
Review
Close interactions between lncRNAs, lipid metabolism and ferroptosis in cancer
1. The Key Laboratory of Model Animal and Stem Cell Biology in Hunan Province, Hunan Normal University, Changsha, 410013 Hunan, China.
2. School of Medicine, Hunan Normal University, Changsha, 410013 Hunan, China.
3. School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, 210013 Jiangsu, China.
4. Department of Head and Neck Surgery, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013 Hunan, P.R. China.
5. Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Department of Pathology, Xiangya Hospital, School of Basic Medicine, Central South University, Changsha, 410078 Hunan, China.
#These authors contributed equally to this work.
Received 2021-8-17; Accepted 2021-10-1; Published 2021-10-25
Abstract
Abnormal lipid metabolism including synthesis, uptake, modification, degradation and transport has been considered a hallmark of malignant tumors and contributes to the supply of substances and energy for rapid cell growth. Meanwhile, abnormal lipid metabolism is also associated with lipid peroxidation, which plays an important role in a newly discovered type of regulated cell death termed ferroptosis. Long noncoding RNAs (lncRNAs) have been proven to be associated with the occurrence and progression of cancer. Growing evidence indicates that lncRNAs are key regulators of abnormal lipid metabolism and ferroptosis in cancer. In this review, we mainly summarized the mechanism by which lncRNAs regulate aberrant lipid metabolism in cancer, illustrated that lipid metabolism can also influence the expression of lncRNAs, and discussed the mechanism by which lncRNAs affect ferroptosis. A comprehensive understanding of the interactions between lncRNAs, lipid metabolism and ferroptosis could help us to develop novel strategies for precise cancer treatment in the future.
Keywords: LncRNAs, Lipid metabolism, Ferroptosis, Cancer
Introduction
During carcinogenesis, cancer cells usually exhibit a series of metabolic abnormalities that support their need for growth and metastasis. In recent years, increasing evidence has shown that lipid metabolism can be reprogrammed in cancer cells [1, 2], which greatly affects the proliferation, invasion and migration of cancer cells. Lipids are hydrophobic molecules that contain thousands of different types of molecules, including cholesterol, fatty acids (FAs), triacylglycerol (TG) and phospholipids (PL) [3, 4]. These compounds play crucial roles in various biological processes, such as the biosynthesis of membrane lipids, energy metabolism, storage and signal transduction [4, 5]. Cancer cells cause dysregulation of lipid metabolism by affecting the synthesis, uptake, modification, degradation, and transport of these lipids in cells, thus affecting their normal physiological function [3, 6]. In addition, a growing body of studies has found that aberrant lipid metabolism is closely related to ferroptosis, a new type of cell death.
Ferroptosis is a novel form of programmed cell death caused by lipid peroxidation (LPO) [7], which is characterized by the accumulation of LPO products induced by lethal reactive oxygen species (ROS) [8]. Polyunsaturated free fatty acids (PUFAs) are rich in cell membranes and organelle membranes and tend to react with ROS and cause cell damage [9]. Since saturated membrane lipids are not sensitive to peroxidation, highly saturated membrane lipids help protect tumor cells from ROS damage. In tumors, tumor cells can express high levels of antioxidant proteins to reduce ROS levels and prevent the destruction of redox homeostasis [10]. Therefore, the discovery of ferroptosis may provide more ways to treat cancer.
Long noncoding RNAs (lncRNAs) are broadly defined as transcripts of more than 200 nucleotides that are not translated into proteins; lncRNAs were historically considered junk DNA and not taken seriously. However, in recent years, it has been increasingly found that lncRNAs have a place in the occurrence and development of cancer [11]. LncRNAs have complex secondary and tertiary structures and diverse subcellular localizations. On the basis of their subcellular localization, lncRNAs can be classified as nuclear, cytoplasmic, and mitochondrial lncRNAs [12]. Functionally, nuclear lncRNAs seem to preferentially play roles in chromatin remodeling, transcriptional regulation and RNA processing, while cytoplasmic lncRNAs can modulate the stability or translation of mRNA and influence the cell signal cascade [13]. Mitochondrial lncRNAs may act as retrograde signaling molecules, coordinating gene expression in the nucleus and mitochondria and ultimately triggering cellular signaling pathways [14]. Many lncRNAs have been regarded as crucial factors in cancer development by affecting a series of cellular processes, including cell proliferation [15, 16], differentiation [17, 18], metastasis [19, 20] and apoptosis [21, 22], at different levels. Therefore, as a regulatory factor, lncRNAs can regulate tumor lipid metabolism and ferroptosis in carcinoma by mediating the expression of enzymes related to lipid metabolism and ferroptosis-related genes.
Therefore, this article will discuss three major points: 1) several abnormal lipid metabolisms (cholesterol, FAs, TG and PL) regulated by lncRNAs; 2) the effect of lipid metabolism on the expression of lncRNAs; and 3) lncRNAs affecting ferroptosis through lipid metabolism. Elaborating the regulatory relationships of these three factors provides new therapeutic targets and ideas for cancer treatment, which may help alleviate the current clinical difficulties in the treatment of cancer.
LncRNAs associated with cholesterol reprogramming in cancer
Cholesterol metabolism is vital for cellular and systemic biological functions. For instance, cholesterol regulates membrane fluidity and permeability, is an essential component in mammalian cell membranes, and is the precursor of bile acid, cholecalciferol and steroid hormones [23]. Dysregulated cholesterol balance not only promotes cardiovascular disease but also increases the risk of occurrence of other diseases, such as neurodegenerative diseases and cancers [24]. Normally, cellular cholesterol metabolism reflects the dynamic balance between synthesis, uptake, storage and efflux [3]. Over the past decade, the cancer research community has witnessed growing interest in cholesterol reprogramming, including the study of how cancer cells reprogram their cholesterol metabolism, how these reprogramming-derived metabolites consequently promote the progression of cancer and the identities of the key regulators in cholesterol reprogramming [25, 26]. LncRNAs can interact with RNA, chromatin and protein, thus modulating mRNA stability, chromatin structure and the function of proteins (including transcription factors), which makes lncRNAs an important group of crucial factors governing cholesterol metabolism processes in cancer [27] (Figure 1 and Table 1).
LncRNAs regulate cholesterol synthesize and uptake
Cholesterol is an indispensable substance in the human body and can be synthesized de novo or ingested from an external source [28]. Acetyl-CoA and NADPH are the basic raw materials for cholesterol synthesis. Acetyl-CoA can be reduced to mevalonate (MVA) under a series of enzymatic reactions with hydroxymethylglutaryl-coenzyme A reductase (HMGCR) as the key enzyme. Then, MVA undergoes decarboxylation and phosphorylation to generate isoprenoids [29]. After the condensation of isoprenoids, 30-carbon squalene is formed. Then, squalene is cyclized to lanosterol under the action of squalene epoxidase (SQLE) [30]. Finally, lanosterol undergoes oxidation, decarboxylation and reduction reactions to generate cholesterol [31]. When the synthesis of cholesterol is insufficient, the low-density lipoprotein receptor (LDLR) can mediate exogenous absorption to maintain the balance of cholesterol in the body [32]. In addition to the increase in cholesterol, which can provide growth requirements for tumor cells, the isoprenoid produced in the MVA pathway can also contribute to the occurrence and development of cancer [33]. Isoprenoids can posttranslationally prenylate small GTP-binding proteins (GTPases), such as the Ras and Rho families, and give them carcinogenic capacity through specific binding to the cell membrane [34]. It is worth mentioning that isopentenyl diphosphate (IPP), the isoprenoid precursor produced in the MCV pathway, can mediate the synthesis of glutathione peroxidase 4 (GPX4) and participate in cholesterol oxidation [35-37]. The hydrogen atom at position C-7 in cholesterol can be taken up by free radicals and then react with diatomic oxygen to produce cholesterol peroxide, leading to lipid peroxidation. However, GPX4 can reduce the occurrence of lipid peroxidation by converting lipid peroxides into lipid alcohols under its antioxidant effect [38, 39]. Therefore, studying the mechanism of the effect of lncRNA on the synthesis and uptake of cholesterol can find ways to inhibit cholesterol reprogramming so that it cannot meet the growth conditions of tumor cells.
It has been reported that lncRNAs can control the expression of HMGCR and LDLR by regulating sterol regulatory element-binding protein 2 (SREBP2) [40]. SREBPs are a class of transcription factors controlling lipid homeostasis by modulating the expression of enzymes required for lipogenesis. SREBP includes three subtypes: SREBP-1a, SREBP-1c, and SREBP-2. It has been indicated that SREBP2 seems preferential to activate the transcription of genes involved in cholesterol synthesis [41-43]. When cholesterol is insufficient, endoplasmic reticulum (ER) cholesterol, as a sensor of intracellular cholesterol homeostasis, can trigger the transport of SREBP2 from the ER to the Golgi apparatus and then to the nucleus to directly regulate the expression of cholesterol-related HMGCR and LDLR [23, 40]. Therefore, directly inhibiting SREBP2 through cholesterol starvation may be an effective strategy against cancer [44]. Yu et al. found that lncRNA SNHG16 is highly expressed in pancreatic cancer and can directly sponge miR-195 to regulate the expression of SREBP2 to accelerate the progression of pancreatic cancer. Moreover, the reduction in pancreatic cancer cell adipogenesis after inhibiting the expression of SREBP2 further proves that lncRNA SNHG16 may fuel the growth of pancreatic cancer cells by regulating the miR-195/SREBP2 axis to provide an energy supply [45].
SQLE is another key controlling enzyme in the MVA pathway. It has been confirmed that SQLE is a metabolic oncogene in breast cancer and is associated with a poor prognosis of breast cancer (BC) [46]. Recent studies have discovered that lnc030 is highly expressed in breast cancer stem cells (BCSCs) and can work with poly(rC)-binding protein 2 (PCBP2) to stabilize the expression of SQLE. PCBP2 is an RNA-binding protein that acts as an intermediary between lnc030 and SQLE. PCBP2 promotes the stability of SQLE mRNA and the production of cholesterol by combining with the lnc030 fragment 301-412 nt and the 3'-UTR of SQLE to form a ternary complex. It has been reported that increased cholesterol synthesis activates PI3K/AKT, the consistent cancer signaling pathway, which is involved in BCSC stemness maintenance [47]. Therefore, lnc030 can promote the occurrence and growth of BC through the SQLE/cholesterol/PI3K/AKT signaling axis.
The known functions of lncRNAs in tumor lipid metabolism. The main lipids in tumor lipid metabolism include cholesterol, FAs, TG and PL. LncRNAs regulate the synthesis and catabolism of these four types of lipids through a variety of mechanisms, thereby participating in the occurrence and development of cancer.
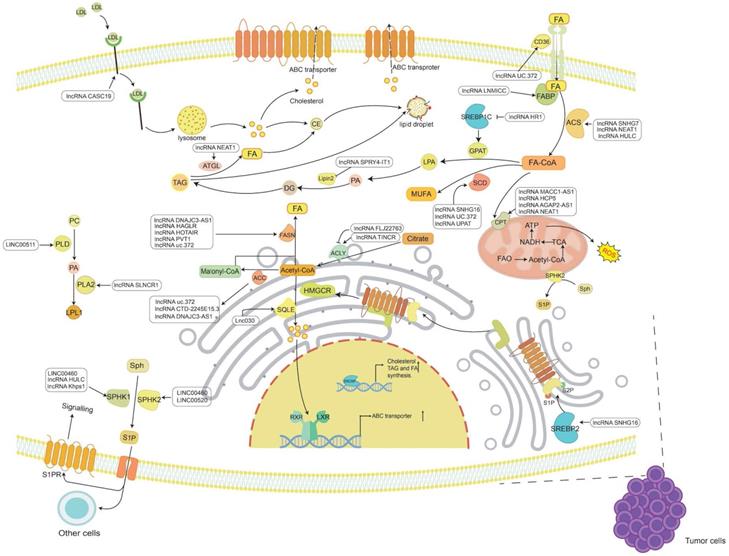
Summary of lipid metabolism-associated lncRNAs in cancer
| LncRNA | Associated type of effect | Target | Influence to target | Cancer type | Reference |
|---|---|---|---|---|---|
| LncRNA SNHG16 | Cholesterol synthesizes | SREBP2 | Up | Pancreatic cancer | [45] |
| Lnc030 | Cholesterol synthesizes | SQLE | Up | Breast cancer | [47] |
| LncRNA CASC19 | Cholesterol uptake | LDLR | Up | Non-small cell lung cancer | [48] |
| LncRNA TINCR | De novo synthesis of fatty acids | ACLY | Up | Nasopharyngeal carcinoma | [77] |
| LncRNA FLJ22763 | De novo synthesis of fatty acids | ACLY | Down | Gastric cancer | [78] |
| LncRNA DNAJC3-AS1 | De novo synthesis of fatty acids | ACC/FASN | Up | Colorectal cancer | [87] |
| LncRNA CTD-2245E15. 3 | De novo synthesis of fatty acids | ACC1 | Up | Non-small cell lung cancer | [85] |
| LncRNA HAGLR | De novo synthesis of fatty acids | FASN | Up | Non-small cell lung cancer | [94] |
| LncRNA HOTAIR | De novo synthesis of fatty acids | FASN | Up | Nasopharyngeal carcinoma | [96] |
| LncRNA PVT1 | De novo synthesis of fatty acids | FASN | Up | Osteosarcoma | [98] |
| LncRNA SNHG16 | De novo synthesis of fatty acids | SCD | Up | Colorectal cancer | [104] |
| LncRNA uc.372 | De novo synthesis of fatty acids | ACC/FASN/SCD1 | Up | Liver cancer | [105] |
| LncRNA UPAT | De novo synthesis of fatty acids | SCD1 | Up | Colon cancer | [107] |
| LncRNA uc.372 | Exogenous uptake of fatty acids | CD36 | Up | Liver cancer | [105] |
| LncRNA LNMICC | Exogenous uptake of fatty acids | FABP5 | Up | Cervical cancer | [120] |
| LncRNA SNHG7 | Oxidation of fatty acids | ACSL1 | Up | Thyroid cancer | [130] |
| LncRNA HULC | Oxidation of fatty acids | ACSL1 | Up | Hepatocellular carcinoma | [132] |
| LncRNA NEAT1 | Oxidation of fatty acids | ACSL4 | Up | Prostate cancer | [131] |
| LncRNA MACC1-AS1 | Oxidation of fatty acids | CPT1 | Up | Gastric cancer | [121] |
| LncRNA HCP5 | Oxidation of fatty acids | CPT1 | Up | Gastric cancer | [142] |
| LncRNA AGAP2-AS1 | Oxidation of fatty acids | CPT1 | Up | Breast cancer | [122] |
| LncRNA NEAT1 | Oxidation of fatty acids | CPT1 | Up | Breast cancer | [144] |
| LncRNA SPRY4-IT1 | Synthesis of triglyceride | Lipin-2 | Down | Melanoma | [150] |
| LncRNA HR1 | Synthesis of triglyceride | SPEBP-1c | Down | Liver cancer | [152] |
| LncRNA NEAT1 | Degradation of triglyceride | ATGL | Up | Hepatocellular carcinoma | [158] |
| LncRNA Khps1 | Phospholipid metabolism | SphK1 | Up | Osteosarcoma | [169] |
| LncRNA HULC | Phospholipid metabolism | SphK1 | Up | Hepatocellular carcinoma | [170] |
| LINC00460 | Phospholipid metabolism | SphK1 | Up | Colorectal cancer | [171] |
| LINC00460 | Phospholipid metabolism | SphK2 | Up | Papillary thyroid carcinoma | [172] |
| LINC00520 | Phospholipid metabolism | SphK2 | Up | Papillary thyroid carcinoma | [173] |
| LINC00511 | Phospholipid metabolism | PLD1 | Up | Cervical cancer | [178] |
| LncRNA SLNCR1 | Phospholipid metabolism | PLA2 | Up | Non-small cell lung cancer | [181] |
| LncRNA HULC | Cholesterol | FOXM1 | Up | Hepatocellular carcinoma | [185] |
| LncRNA UCA1 | Palmitic acid | ERK/MMP-9 | Up | Gastric cancer | [190-192] |
| LncRNA ANRIL | Phospholipid | / | / | Large cell lung cancer | [193] |
In addition, lncRNAs can also directly affect the expression of LDLR to promote tumor development. Wang et al. found that lncRNA CASC19 was significantly upregulated in non-small cell lung cancer (NSCLC) and was positively correlated with the proliferation and metastasis of NSCLC cells. It was predicted and proved that LDLR is the downstream target gene of miR-301b-3p in NSCLC, and it is inhibited when interacting with miR-301b-3p. However, CASC19 can sponge miR-301b-3p and restore the effect of LDLR. Therefore, CASC19 plays a role in NSCLC by targeting the miR-301b-3p/LDLR axis to promote the proliferation and metastasis of cancer cells [48].
LncRNAs regulate cholesterol efflux
Most peripheral cells and tissues lack the ability to metabolize cholesterol [49]. When the cholesterol in the cell exceeds what the cell needs, cholesterol is either converted to cholesterol ester (CE) by the action of cholesterol acyltransferase (ACAT) [50], which is stored in lipid droplets or secreted in lipoproteins, or is excreted from the cell via an ATP binding cassette (ABC) transporter. Liver X receptors (LXRs) can be used as cholesterol sensors [51]. When intracellular cholesterol is at a high level, LXRs are activated by cholesterol derivative oxysterols [52], and then they can bind to retinoid X receptor alpha (RXRα) as an obligate heterodimerization partner to direct repeat 4 (DR4) in the promoter of ABC transporters [53, 54], thereby activating the expression of ABC transporters to promote cholesterol export.
LXR is composed of alpha and beta subtypes. A number of studies have shown that the activation of LXR can significantly inhibit tumor progression. For example, activating the expression of LXR can inhibit the protein associated with cell proliferation in BC [55]. Activating LXR can block the G1 phase of cancer cells in intestinal tumors and increase caspase-dependent apoptosis to inhibit the development of cancer [56]. In addition, LXR-623, as an activator of LXR, can effectively inhibit the development of glioblastoma (GBM) by activating LXRβ, lowering cholesterol levels, and inducing apoptosis [57]. ATP binding box transporter A1 (ABCA1), a subtype of ABC, can be activated by LXRs to promote cholesterol efflux [58]. When ABCA1 expression is reduced, cholesterol is allowed to accumulate in cells. Cholesterol accumulation leads to reduced membrane fluidity and inhibition of the mitochondrial permeability transition (MPT), which prevents the release of cell death-promoting molecules in mitochondria [59], ultimately promoting the progression of cancer. However, the current research on the regulatory mechanism of lncRNAs on ABC is mostly limited to noncancer diseases. For example, lncRNA GAS5 is highly expressed in atherosclerosis and can recruit enhancer of zeste homolog 2 (EZH2) to the ABCA1 promoter region to promote histone methylation modification of ABCA1. Eventually, transcription of ABCA1 is inhibited, leading to cholesterol accumulation and atherosclerosis [60]. In addition, lncRNA ENST00000602558.1 can directly bind to P65 to promote P65 binding to the promoter of ABCG1 and inhibit the expression of ABCG1 mRNA and protein in vascular smooth muscle cells. The reduction in cholesterol efflux mediated by ABCG1 further leads to dyslipidemia and atherosclerosis [61].
LncRNAs mediate abnormal fatty acid metabolism in cancer
Many lipids are synthesized from FAs, a class of molecules consisting of hydrocarbon chains of varying lengths and degrees of desaturation [6]. Within cells, FAs have various functions, including being a component of the membrane, conducting signals, or being oxidized to release energy. FAs are either obtained from an external source or synthesized from scratch. Under normal conditions, most normal cells preferentially use exogenous FAs to meet their lipid requirements [62]. FAs must be activated by covalent modification of fatty acyl-CoA synthetase (ACS) to synthesize lipids or decompose to produce energy [63]. In cancer, FA metabolism is undoubtedly altered due to the high energy requirements of rapidly proliferating cells [6, 64, 65]. Therefore, the roles of lncRNAs in FA uptake, de novo synthesis, and oxidative degradation in tumor progression are also worthy of study and discussion (Figure 1 and Table 1).
LncRNAs regulate the de novo synthesis of fatty acids
Most normal cells have the ability to take up lipids from the extracellular environment and are thus more inclined to take up FA exogenously[62, 66]. In contrast, an increase in the demand for lipids by tumor cells has been observed in tumors, such as signaling molecules and membrane biosynthesis. Therefore, the de novo synthesis of FA plays a dominant role in tumors [67-69]. The increased demand for FA synthesis becomes an adaptation to the high metabolic demand of cancer cells. The de novo synthesis of FAs requires the action of a variety of enzymes. First, glucose is decomposed into pyruvate and then enters the mitochondria to oxidize and decarboxylate to generate acetyl-CoA; since acetyl-CoA cannot directly penetrate the mitochondrial membrane and needs to synthesize FAs in the cytoplasm, it must first condense with oxaloacetate to form citric acid and then enter the cytoplasm. Then, acetyl-CoA is generated under the action of ATP citrate lyase (ACLY), and finally, saturated FAs are synthesized under the continuous condensation reaction of acetyl-CoA carboxylase (ACC) carboxylation and fatty acid synthase (FASN) [64, 70]. Additionally, saturated FAs can be desaturated by stearoyl-CoA desaturase (SCD) according to cellular requirements to produce monounsaturated FAs [64]. Therefore, inhibiting these enzymes and reducing FA synthesis will effectively limit the growth of cancer cells.
ATP citrate lyase
ATP citrate lyase (ACLY) is an enzyme that catalyzes an important step in FA synthesis. It can catalyze the conversion of citric acid and coenzyme A (CoA) to acetyl-CoA and oxaloacetic acid. Acetyl-CoA is essential for FA synthesis and cancer cell proliferation. ACLY is abnormally expressed and active in many tumors [71, 72], such as renal carcinoma [73], pancreatic cancer [74], BC [75] and gastric cancer (GC) [76].
Zheng et al. found that lncRNA TINCR was highly expressed in nasopharyngeal carcinoma (NPC) cells. RNA pull-down assay and RNA-EMSA analysis confirmed that the 1-876 nt region of TINCR can bind ACLY. Subsequently, it was shown that TINCR can inhibit the ubiquitin-mediated degradation of ACLY to protect the proteasome-dependent degradation of ACLY, thereby maintaining the stability of the ACLY protein. The increased stability of the ACLY protein increases the level of acetyl-CoA, hence increasing the content of free fatty acids (FFAs), leading to the increased proliferation, metastasis and chemoresistance of cancer cells [77]. Zhang et al. demonstrated that lncRNA FLJ22763 is downregulated in GC tissue and is closely related to the survival of patients. The researchers revealed a significant negative correlation between FLJ22763 and ACLY mRNAs in GC. After FLJ22763 overexpression, ACLY was significantly downregulated, hence inhibiting GC cell malignancy and xenograft tumor growth [78]. In addition, in recent years, researchers have used the statistical method of weighted correlation network analysis (WGCNA) to determine the coexpression network of lncRNAs and ACLY in GC. WGCNA can identify the genes most related to cancer in the database. Then, a series of methods, including univariate and multivariate Cox regression analyses and Pearson correlation and hypergeometric tests, confirmed that ACLY was correlated with lncRNA PPP1R26-AS1, lncRNA DLEU1 and lncRNA TMPO-AS1 [79]. However, further experiments are needed to verify the relationship between these lncRNAs and ACLY.
In addition, some lncRNAs that regulate the expression of ACLY are also associated with viral infections. Lipid production has a positive effect on virus replication [80]. Linc-Pint is significantly downregulated in HCV-infected hepatocytes. Linc-Pint overexpression may downregulate the protein level of serine/arginine protein-specific kinase 2 (SRPK2) through proteasome degradation [81]. SRPK2 activates serine/arginine (SR) proteins involved in mRNA splicing and maturation [82]. It has been reported that SRPK2 can promote effective splicing by the phosphorylation of SR proteins to increase the stability of ACLY and FASN mRNA [83]. Therefore, the overexpression of Linc-Pint decreased the expression of SRPK2, which further reduce the stability of ACLY and FASN mRNA. Finally, the HCV-induced lipid production pathway is inhibited to limit the replication of HCV [81].
Acetyl-CoA carboxylase
Acetyl-CoA can be carboxylated to malonyl-CoA under the action of acetyl-CoA carboxylase (ACC), which is an indispensable step in FA synthesis [3]. ACC has two different subtypes, ACC1 and ACC2, and the functions of malonyl-CoA produced by them are different. The malonyl-CoA produced under the action of ACC1 mainly promotes the synthesis of FAs, while the malonyl-CoA produced under the action of ACC2 mainly inhibits the oxidation of FAs [84], keeping the FA content in the body at a high level. To further meet the needs of tumor cells, ACC is highly expressed in NSCLC [85], cervical squamous cell carcinoma [86] and colorectal cancer [87].
The mechanisms by which lncRNAs regulate ACC in different tumors are also different. LncRNA DNAJC3-AS1 is highly expressed in colorectal cancer and can regulate the expression of ACC and FASN by activating PI3K/AKT, a recognized oncogenic pathway, thereby promoting tumor progression [87]. Recent studies have demonstrated that lncRNA CTD-2245E15.3 is highly expressed in NSCLC, and its inhibition can regulate lipid metabolism-related genes. Based on this conclusion, the study also showed that the knockout of CTD-2245E15.3 can phosphorylate the Ser117 site of ACC1. The Ser117 site of ACC1 is the inhibitory site of enzyme activity. Its phosphorylation inhibits the enzyme activity of ACC1, which further leads to lipid synthesis obstacles and ultimately inhibits tumor growth. It is concluded that CTD-2245E15.3 promotes the progression of non-small cell lung cancer by regulating the enzyme activity of ACC1 [85].
Fatty acid synthase
Fatty acid synthase (FASN) is a multifunctional enzyme that catalyzes the biosynthesis of palmitate esters in an NADPH-dependent manner and thus participates in the synthesis of FAs [88]. FASN is widely expressed in normal cells, and its promotion of expression leads to an increase in FA synthesis. An imbalance in FASN expression can cause many diseases. For example, the high expression of FASN has been demonstrated to be closely related to poor prognosis in various cancers, such as BC [89] and NSCLC [90].
HAGLR is a lncRNA transcribed from the HOXD cluster on human chromosome 2 that is upregulated in multiple cancers, including colon cancer [91], hepatocellular carcinoma [92] and BC [93], and is closely related to progression and unfavorable prognosis. Lu et al. discovered that the level of HAGLR expression in NSCLC increased and was associated with poor prognosis in patients. The level of FASN is positively correlated with the expression of HAGLR. The expression of FASN in NSCLC decreased with the knockdown of HAGLR, which reduced the FFA content in cells and inhibited the proliferation, invasion and tumorigenesis of NSCLC cells [94]. In addition, lncRNA HOTAIR plays a role in regulating chromatin dynamics in gene regulation. As a proto-oncogene, it is highly expressed in a variety of cancers [95]. Knockout of HOTAIR can reduce FASN expression and inhibit FA synthesis in NPC cells, thereby inhibiting their proliferation and invasion [96]. It is worth mentioning that matrix metalloproteinase-9 (MMP-9), as a potential cancer marker [97], has been downregulated in the knockout of HAGLR and HOTAIR [94, 96]. Therefore, whether FASN affects MMP-9 plays a role and needs further verification. Moreover, Zhou et al. found that lncRNA PVT1 is overexpressed in osteosarcoma, reducing the survival rate of patients with osteosarcoma. PVT1 mainly acts as a competitive endogenous RNA (ceRNA) to negatively regulate miR-195 in osteosarcoma cells to enhance FASN expression, thereby promoting osteosarcoma cell migration and invasion [98].
Stearoyl-CoA desaturase
SCD is a major enzyme involved in the synthesis of monounsaturated FAs. Two types of SCD isoforms, SCD1 and SCD5, have been found in humans [99]. Among them, SCD1 is widely expressed in tissues. The ratio between saturated FAs and unsaturated FAs is very important for cancer cells, and its changes will affect cell fluidity and protein dynamics [84]. The increase in unsaturated FAs not only facilitates the proliferation and metastasis of cancer cells but also inhibits cell apoptosis. In many types of cancers, increased expression of SCD1 can lead to the proliferation and invasion of cancer cells, and inhibition of SCD1 expression can inhibit tumor progression in vivo, such as prostate cancer [100], bladder cancer [101], lung cancer [102] and clear cell kidney cell carcinoma [103].
The expression of lncRNA SNHG16 is upregulated in colorectal cancer and is regulated by c-Myc. The upregulation of SNHG16 was confirmed after c-Myc overexpression. To further clarify the potential molecular mechanism of SNHG16, genome-wide transcription profiling was performed after knocking down SNHG16. Next, Ingenuity Pathway Analysis (IPA) was used to determine that knocking out SNHG16 can affect the expression of lipid metabolism-related genes. The study discovered that the miRNA bound by SNHG16 cotargeted the 3'-UTR of SCD mRNA. Therefore, SNHG16 may upregulate the expression of SCD through a ceRNA mechanism to promote the proliferation and migration of colorectal cancer cells [104]. In addition, UC (ultraconserved, UC) RNA is also a long noncoding RNA. After overexpression of uc.372 in liver cancer HepG2 cells, FASN, ACC, SCD1 and CD36 were all upregulated. To further study the molecular mechanism of uc.372 regulating the expression of ACC, FAS, SCD1 and CD36. Microarray analysis showed that pri-miR-195 and pri-miR-4668 were complementary to the ultraconserved region of uc.372. Overexpression of uc.372 can inhibit the maturation of miR-195 and miR-4668 by specifically binding to pri-miR-195 and pri-miR-4668 [105]. Based on previous research, ACC and FAS are the target genes of miR-195 [106]. Experimental prediction and verification proved that SCD1 and CD36 are the target genes of miR-4668 [105]. The overexpression of uc.372 reduced the suppression of ACC and the expression of FASN by miR-195 and the suppression of SCD1 and the expression of CD36 by miR4668. Therefore, it can be concluded that uc.372 can upregulate ACC, FASN, SCD1 and CD36 through the pri-miR-195/miR-195 and pri-miR-4668/miR-4668 signal axes to drive fat accumulation in HepG2 cells [105]. Moreover, lncRNA UPAT is necessary for the tumorigenicity of colon cancer cells. Through a series of experiments to identify proteins that may be related to UPAT, researchers found that UPAT can interfere with the ubiquitination and degradation of UHRF1 in colon tumors through the proteasome, thereby maintaining the stability of UHRF1. Knockout of UPAT or UHRF1 reduces the expression of SCD1, but UHRF1 has nothing to do with the SCD1 promoter region. Therefore, it is speculated that SCD1 is not a direct target of UHRF1 but is indirectly upregulated downstream of UHRF1 and UPAT. The effect of lncRNA UPAT on SCD1 needs further study [107].
LncRNA regulates the exogenous uptake of fatty acids
Although the de novo synthesis of FA in cancer occupies a major position in the synthesis of macromolecules, some FAs can also be transported by certain proteins [64], and circulating FAs can be absorbed and utilized to provide support for the survival and development of cancer cells [84]. Therefore, transport proteins such as fatty acid translocase (FAT/CD36) and fatty acid-binding protein (FATP) in the FA uptake pathway may become potential targets for cancer treatment.
Fatty acid translocase
FAT/CD36 is a widely expressed transmembrane protein that mediates the uptake of FAs. It can also bind to carnitine palmityl transferase 1 (CPT1) in fatty acid oxidation (FAO), thus promoting FAO and providing sufficient energy for the rapid proliferation and development of cancer cells [108]. However, few studies have demonstrated the influence of noncoding RNAs on FAT/CD36 regulation in cancer. Only studies have shown the effect of noncoding RNAs on FAT/CD36 in atherosclerosis.
CD36 is a key mediator of macrophage phagocytosis of oxidized low-density lipoprotein (oxLDL) in atherosclerosis [109]. In recent years, studies have found that macrophages can change their phenotypes according to changes in the microenvironment and thus have diverse functions. The two main macrophage phenotypes are classically activated macrophages (M1) and selectively activated macrophages (M2) [110]. The M1 phenotype has obvious proinflammatory and antitumor activities, while the M2 phenotype is an anti-inflammatory phenotype with protumor activity [111]. Although both M1 and M2 macrophages are present in atherosclerosis, M1 macrophages are a dominant phenotype associated with plaque progression [112]. M1 macrophages form foam cells after phagocytosing oxLDL. Foam cells then secrete proinflammatory mediators to further aggravate the production of unstable atherosclerotic plaques [113]. LncRNA MALAT1 can accumulate β-catenin on the CD36 promoter-binding site in macrophages, promote CD36 transcription and lipid uptake [114], and ultimately accelerate the occurrence of atherosclerosis. LncRNA PELATON is rich in unstable atherosclerosis, especially in the areas where the nuclei of macrophages gather. RNA sequencing found that there was a strong positive correlation between PELATON and CD36. With the decrease in PELATON, CD36 expression was significantly reduced, leading to marked reductions in macrophage phagocytosis and lipid absorption, which inhibited plaque progression [115]. Apart from atherosclerosis, macrophages are also closely related to the poor prognosis of tumors [116]. Tumor-associated macrophages (TAMs) are the main component of inflammatory cells that infiltrate cancer. In advanced cancers, most macrophages are of the M2 phenotype, which highly stimulates tumor progression [116]. Therefore, whether lncRNAs affect the role of TAMs by regulating CD36 still needs to be studied.
Fatty acid-binding protein
Fatty acid-binding protein (FABP) is involved in FA transport and metabolism, and FABP can bind to FFAs and transport them to various organelles for further oxidation or esterification. Multiple studies have shown that FABP5 can be involved in the occurrence and development of hepatocellular carcinoma (HCC) [117], BC [118] and prostatic carcinomas [119].
Shang et al. found that lncRNA LNMICC was a valuable prognostic predictor of cervical cancer. Multiple regression analysis revealed that high LNMICC expression was significantly related to the BMI of cervical cancer patients. Therefore, it is speculated that LNMICC is related to the reprogramming of FA metabolism in cervical cancer. For further confirmation, the levels of cell-related lipids and the expression of key FA metabolism were measured at different expression levels of LNMICC. Researchers have revealed that the levels of cell-related lipids and the expression of key enzymes for FA metabolism in cervical cancers with different LNMICC levels are different. Next, the researchers further studied how LNMICC exerts its biological function to reprogram FA metabolism. Based on bioinformatics analysis, LNMICC is located upstream of the FABP5 gene, and the mRNA levels of LNMICC and FABP5 are positively correlated. A series of experiments discovered that LNMICC can target the FABP5 promoter region by recruiting NPM1 to interact directly with LNMICC, thereby enhancing the transcription of FABP5. Finally, combined in vivo and in vitro biological function experiments show that LNMICC can promote lymph node metastasis and epithelial-mesenchymal transition (EMT) by directly regulating FABP5-mediated FA metabolic reprogramming [120].
LncRNAs regulate the oxidation of fatty acids
Fatty acid oxidation (FAO) is an important catabolic process in which living organisms use FAs as energy sources. FAs must be activated before oxidation. FAs are activated under the action of ACS to generate acyl-CoA. Acyl-CoA enters the mitochondria under the action of carnitine palmityl transferase (CPT) and is oxidized and decomposed to produce acetyl-CoA, FADH2 and NADH. Acetyl-CoA enters the TCA cycle and is completely oxidized to produce ATP, while NADH and FADH2 enter the electron transport chain to produce ATP. According to reports, many types of cancers show high FAO activity, such as GC [121], BC [122], glioma [123] and acute myeloid leukemia (AML) [124]. FAO can promote tumor development by increasing the production of ATP. NADPH produced by acetyl-CoA in the TCA cycle provides cancer cells with a redox ability to resist oxidative stress [125]. Therefore, targeting key enzymes in FAO may hopefully provide an effective method for cancer treatment.
Acyl-CoA synthetase long chain family member
FAs can be catabolized to acetyl-CoA to promote the production of ATP or serve as the raw material for the synthesis of TAG, PL and CE [63, 126]. However, these two distinct pathways require a common initial step, which is referred to as FA activation. ACS is an enzyme necessary for the activation of FAs. Among them, acyl-CoA synthetase long-chain family members (ACSLs) are the most critical enzymes responsible for the activation of the most abundant long-chain FA metabolism in mammalian cells. ACSLs include ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6 [127]. Different members of ACSLs play various roles in cancer, and their expression varies in different cancers. For example, ACSL4 is highly expressed in colon adenocarcinoma [128], but it inhibits tumors in GC [129].
According to reports, lncRNAs also have the potential to regulate ACSLs. For example, lncRNA SNHG7 plays a carcinogenic effect in thyroid cancer (TC). After SNHG7 is knocked down, the mRNA and protein levels of ACSL1 are suppressed. Cell proliferation and migration experiments revealed that the increase in ACSL1 can reverse the inhibitory effects of SNHG7 on cell proliferation and migration when SNHG7 is depleted. In addition, bioinformatics analysis found that miR-449a can act as a miRNA that simultaneously binds SNHG7 and ACSL1. To analyze the role of miR-499a in the regulation of ACSL1 by SNHG7, researchers conducted RIP and luciferase reporter gene analyses. The experiment concluded that SNHG7 can be used as a sponge for miR-449a, thereby increasing ACSL1 in TC cells and promoting the proliferation and migration of TC cells [130]. Additionally, lncRNA NEAT1 is highly expressed in docetaxel-resistant prostate cancer patients and cell lines. The CCK-8 experiment confirmed that after knocking out NEAT1, the IC50 value of docetaxel on prostate cancer cells decreased significantly. Compared with docetaxel-treated parental cells, NEAT1 knockdown promoted the sensitivity of cells to docetaxel and reduced cell proliferation and invasion. To study the potential molecular mechanism of NEAT1 in prostate cancer, bioinformatics predictions and experiments proved that miR-34a-5p and miR-204-5p are potential targets of NEAT1 and are inhibited by NEAT1. Next, the downstream targets of miR-34a-5p and miR-204-5p were predicted and verified to be ACSL4. miR-34a-5p and miR-204-5p can inhibit the expression of ACSL4 by targeting the 3'-UTR of ACSL4 and then reduce the ability of prostate cancer cells to be resistant to docetaxel. In summary, NEAT1 can enhance the expression of ACSL4 by sponging miR-34a-5p and miR-204-5p, thereby promoting docetaxel resistance in prostate cancer cells and accelerating the progression of prostate cancer [131]. In addition, studies have shown that lncRNA HULC can regulate abnormal lipid metabolism in the body through the miR-9/PPARA/ACSL1/signaling pathway [132], increase the accumulation of TG and CE in HCC tissues, and promote the growth and development of HCC. The mechanism of this signaling pathway will be explained in detail below.
Carnitine palmityl transferase
At present, it is known that FAO plays a role in inducing cancer cell metastasis and chemotherapy resistance and improving the stemness of cancer cells [65, 121, 133-135]. Among them, CPT1, as the rate-limiting enzyme of FAO, provides the first and rate-limiting steps of FA transport to mitochondria for oxidation. CPT1 can directly control the production of ATP and NADPH in the FAO process, which constitutes an important part of cancer metabolic adaptation [125]. Therefore, fully exploring the molecular mechanism of lncRNA regulating CPTI can identify a new therapeutic window in cancer treatment intervention.
Metastasis-associated in colon cancer-1 (MACC1) is a transcriptional regulator of EMT. MACC1 is upregulated in various tumors and enhances cell proliferation, invasion and chemotherapy resistance [136-140]. MACC1-AS1 is the cognate antisense lncRNA of MACC1. Recent studies have shown that MACC1-AS1 can be induced to be expressed by mesenchymal stem cells (MSCs) [121]. In previous reports, MSCs secreted TGF-β1, a key cytokine. TGF-β exerts cellular effects by binding to TGFβ receptors (TGFβR-I and TGFβR-II) and then activates the downstream SMAD family [141]. In a recent study, it was found that when MSCs are cocultured with GC cell lines, TGF-β1 secreted by MSCs can be directly combined with TGFβR-I and TGFβR-II in GC cells to activate SMAD2 and SMAD3 and then induce the upregulation of MACC1-AS1 in GC cells. The upregulation of MACC1-AS1 activates the FAO pathway in GC cells, and the expression levels of FAO-related enzymes (CPT1 and ACS) are significantly increased, which promotes stemness and chemoresistance in GC. In addition, to clarify the potential mechanism of MACC1-AS1 on FAO-dependent dryness and chemical resistance, it was found that miR-145-5p is located downstream of MACC1-AS1 based on the prediction of the LncRNASNP database and experimental verification, and MACC1-AS1 can directly bind to miR-145-5p and inhibit the expression of miR-145-5p. miR-145-5p partially reversed the promoting effect of MACC1-AS1 on the expression of CPT1 and stem genes and inhibited the occurrence of drug resistance and FAO [121]. In summary, the TGF-β1/MACC1-AS1/miR-145-5p/CPT1 signaling axis contributes to FAO-dependent stem and chemoresistance, indicating that targeting this signaling pathway may be a potential strategy to inhibit stem cells and chemoresistance induced by MSCs. LncRNAs termed HCP5 could be induced in GC cells by coculture with MSCs and were reported to play an important role in elevating GC cancer cell stem properties and chemoresistance and in predicting a poor prognosis. The pull-down assay showed that miR-3619-5p can be pulled down by HCP5 in GC cells, indicating that there is an interaction between HCP5 and miR-3619-5p. Then, in the biological function experiment, the effects of HCP5 and miR-3619-5p were opposite. HCP5 can exert a sponge effect on miR-3619-5p to promote the activity of CPT1 in GC cells. To further explore the molecular mechanism of this, KEGG analysis revealed that miR-3619-5p was significantly related to the AMPK pathway. The most significantly downregulated gene after miR-3619-5p overexpression was PPARG coactivator 1α (PPARGC1A), a key regulator of the AMPK pathway [142]. It was previously reported that PGC1α, the protein product of PPARGC1A, is a transcriptional coactivator responsible for lipid metabolism [143]. This study revealed that PGC1α can form a transcription complex with CEBPB to activate the transcription of CPT1 and ultimately promote FAO in GC cells. Therefore, the FAO of GC cells can be driven by the HCP5/miR-3619-5p/PPARGC1A/PGC1α/CPT1 axis in GC cells activated by MSCs and ultimately promote the chemoresistance and stemness of GC cells [142].
In addition, another lncRNA induced by MSCs is lncRNA AGAP2-AS1, which was significantly upregulated in BC cells cocultured with MSCs. Bioinformatics analysis showed that the target gene of AGAP2-AS1 was enriched in FAO metabolism. Then, experiments verified that AGAP2-AS1 mediated stemness and trastuzumab resistance by targeting CPT1 and ACS. Further research on the regulatory mechanism of AGAP2-AS1 found that the interaction between AGAP2-AS1 and CPT1 is achieved in two ways. First, AGAP2-AS1 can interact with Hur to produce AGAP2-AS1-Hur and then directly bind to CPT1 mRNA to improve the stability of CPT1. Second, AGAP2-AS1 inhibited the expression of miR-15a-5p through sponge action to increase CPT1 mRNA. Under the combined actions of these two pathways, AGAP2-AS1 can upregulate the expression of CPT1 at the mRNA level, and the FAO of BC cells can be promoted, thereby mediating the characteristics of cancer stem cells and the resistance of trastuzumab [122]. In addition, it has been reported that lncRNA NEAT1 can competitively bind to miR-107 in BC cells, indirectly inhibit the inhibitory effect of miR-107 on CPT1, promote FA oxidation, and provide ATP to regulate the growth and metastasis of BC [144].
LncRNAs are closely related to abnormal triglyceride metabolism in cancer
TG is a key energy source composed of FFA. When there is too much FA, most of the FA can be connected to the glycerol backbone to form TG, which is then stored in cytoplasmic lipid droplets (LDs) [145]. This storage itself is not disease-causing, but TG, as a substance for storing FA, can release FA at any time. This ready availability may provide cancer with much energy and promote tumor proliferation and metastasis [64]. The accumulation of lipids can also cause lipotoxicity, inducing apoptosis [146]. Therefore, increasing FA storage and inhibiting FA release may also be cancer suppression strategies [147] (Figure 1 and Table 1).
LncRNAs regulate the synthesis of triglyceride
First, FA is activated into fatty acyl-CoA (FA-CoA), and FA-CoA forms lysophosphatidic acid (LPA) under the action of glycerol 3-phosphate acyltransferase (GPAT). LPA is converted into phosphatidic acid (PA) under the action of acylglycerophosphate acyltransferase (AGPAT). Then, phosphatidic acid phosphohydrolase (PAP or lipin) removes the phosphate group from PA to form diacylglycerol (DG). Finally, diacylglycerol acyltransferase (DGAT) esterifies DG and FA-CoA to TG [148, 149]. Therefore, by reducing FA utilization and increasing its storage as TG, tumor progression can be inhibited. For instance, lncRNA SPRY4-IT1 is highly expressed in melanoma, and the protein related to SPRY4-IT1 is identified as lipin-2 by mass spectrometry analysis. Lipin-2 can be used as the direct target of lncRNA SPRY4-IT1. Downregulation of SPRY4-IT1 resulted in the enhancement of lipin-2 mRNA, protein and enzyme activities. Due to the effective conversion of DAG mediated by lipin-2 to TAG, both the DGAT mRNA expression level and the TG content increased. Based on these results, it is speculated that SPRY4-IT1 knockdown may induce the apoptosis of melanoma cells through lipin-2-mediated lipotoxicity, but this speculation needs further experimental verification [150].
In addition to directly regulating the enzymes in the TG synthesis pathway, lncRNAs can also indirectly regulate the synthesis of FA and TG by regulating SREBP-1c to achieve lipid metabolism reprogramming. SREBP-1c mainly activates the transcription of multiple genes, such as ACLY, ACC, FAS, SCD-1, and GPAT. The enzymes encoded by these genes can participate in the synthesis of FAs and TG [151]. Therefore, targeting SREBP-1c can effectively inhibit the production of lipids and prevent the proliferation of cancer cells. Li et al. found that overexpression of lncRNA HR1 in Huh7 liver cancer cells can inhibit the phosphorylation of AKT. AKT acts as an upstream regulator of Forkhead Box O1 (FoxO1), and inhibition of AKT phosphorylation reduces the nuclear translocation of FoxO1, which causes a large accumulation of FoxO1 in the nucleus [152]. In previous reports, FoxO1 was generally regarded as a tumor suppressor that can antagonize the combination of the LXRα/RXR heterodimer and LXR elements (LXREs) in the promoter region of SPEBP-1c, thereby downregulating the transcription of SREBP-1c [153]. In summary, lncHR1 may inhibit the accumulation of TG in liver cancer cells through the AKT/FoxO1/SREBP-1c pathway and ultimately may reduce the energy supply of cancer cells to inhibit tumor progression [152].
LncRNA regulates the degradation of triglyceride
FA can be stored in the form of TG. When needed, each TG molecule can be sequentially catalyzed by triglyceride lipase (ATGL), hormone-sensitive lipase (HSL) and monoacylglycerol lipase (MAGL) to release three FAs. ATGL is usually considered the key enzyme for the release of FAs from TG stores [154, 155]. High levels of ATGL have been discovered in a variety of cancers, such as BC [156] and lung cancer [157]. Liu et al. found that the expression of ATGL in HCC tissues is upregulated, which is related to poor prognosis. Co-lncRNA software screening and assay identification showed that lncRNA NEAT1 and ATGL were positively correlated in HCC tissues. To explore the molecular mechanism of NEAT1/ATGL, researchers used a series of bioinformatics methods to determine whether there is a potential miRNA to regulate ATGL. Studies have found that miR-124-3p can directly bind ATGL to inhibit the expression of ATGL. However, the interaction between NEAT1 and ATGL may occupy the binding site of miRNA-124-3p and ATGL, weakening the inhibitory effect of miR-124-3p on ATGL [158]. In addition, it has been proven that FA is the main physiological ligand that activates the known oncogene PPARα in liver cancer [159]. NEAT1 abnormally regulates lipolysis, which leads to an increase in FA, which eventually increases the expression of PPARα, thereby promoting the growth and reproduction of HCC cells [158]. These results indicate that NEAT1 can mediate the growth of HCC cells through miR-124-3p/ATGL/DAG + FA/PPARα signaling.
LncRNAs control phospholipid metabolism in cancer
Phospholipids are the key components of cell membranes and can be divided into two categories: glycerophospholipids and sphingomyelin. Phospholipids play important roles in cells, such as chemical energy storage, cell signal transmission, and cell-cell interactions [160]. Since uncontrolled cell proliferation in cancer requires sufficient energy and cell structural units, the phospholipids in cancer cells need to be actively biosynthesized so that phospholipids can perform the abovementioned functions and participate in the development of cancer (Figure 1 and Table 1).
Sphingosine kinases
Sphingosine kinases (SphKs) are key regulatory enzymes that catalyze the formation of sphingosine-1-phosphate (S1P). SphK1 is a cytoplasmic protein. Once Sphk1 is activated by various extracellular signal transduction pathways, it is phosphorylated by ERK1/2 and transferred to the plasma membrane to phosphorylate sphingosine to form S1P [161]. Conversely, SphK2 is mainly located in the nucleus and part of the mitochondria, and S1P can also be produced at these sites [162, 163]. Currently, S1P is involved in regulating cell growth, proliferation, survival, migration and angiogenesis [164]. Therefore, SphKs may be an important tumorigenesis regulator. Further understanding of the molecular mechanisms regulating SphK expression may reveal new directions for therapeutic strategies. It has been found that the abnormal expression of SphKs is associated with the development and poor prognosis of many cancers [165-168]. There are a variety of lncRNAs that can participate in various types of cancer-related biological abnormalities by regulating the expression of SphKs.
In osteosarcoma, Anna et al. found that lncRNA Khps1 is an antisense RNA that can activate the transcription of SphK1. Therefore, researchers have studied the molecular mechanism by which Khps1 regulates the expression of SphK1. SphK1 contains two putative triplex-forming regions (TFRs) upstream of the transcription start site of its subtype B (SphK1-B). Khps1 can directly interact with TFR2 to form a purine-rich RNA-DNA triplet through Hoogsteen hydrogen bond base pairing, which is anchored to the SphK1-B promoter. Khps1 linked to SphK1-B can interact with histone acetyltransferase p300/CBP, which modifies histone acetylation to create an open chromatin structure. The change in chromatin structure promotes the binding of the SphK1-B upstream transcription factor E2F1 to the SphK1-B promoter to activate SphK1 transcription. Interestingly, the DNA fragment containing the Khps1 promoter also has a binding site for the transcription factor E2F1. Combined with kinetic data analysis, E2F1 induces Khps1 transcription before promoting SphK1-B mRNA upregulation. Therefore, elevated levels of E2F1 will induce the transcription of Khps1. Then, Khps1 changes the chromatin structure of SphK1-B to promote the combination of E2F1 and the SphK1-B promoter to activate SphK1-B transcription and ultimately promote the proliferation of osteosarcoma cells and limit cell apoptosis [169]. In HCC, Zhan et al. found that lncRNA HULC can increase the expression levels of SphK1 mRNA and protein and promote the angiogenesis of HCC. Studies on the transcriptional regulation of SphK1 by HULC showed that E2F1 was involved in the activation of SphK1 transcription by HULC. To study the relationship between HULC and E2F1, the 3'UTR of E2F1 mRNA was analyzed. Studies have demonstrated that miR-107 inhibits the expression of E2F1 by targeting the 3'UTR of E2F1 mRNA. Based on the fact that lncRNAs can be used as ceRNAs to regulate miRNAs, experiments have observed that HULC can act as a sponge to inhibit miR-107 to upregulate E21F, thereby promoting the expression of Sphk1. Therefore, these results indicate that HULC sponges miR-107 and activates the SphK1 promoter under the action of the transcription factor E2F1, induces the upregulation of SphK1, and finally stimulates the angiogenesis of HCC [170]. In addition, LINC00460 is significantly upregulated in colorectal cancer. miRcode software was used to predict the potential target of LINC00460. The results show that miR-613 is a direct potential target of LINC00460. LINC00460 can be used as a ceRNA sponge for miR-613 to increase the expression of the downstream target gene Sphk1 of miR-613, thereby promoting the proliferation, migration and invasion of colorectal cancer cells [171].
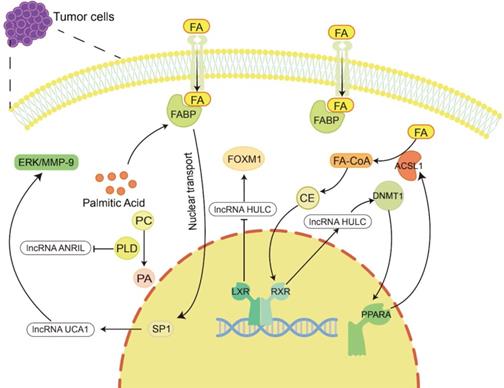
Lipid metabolism regulates the occurrence and development of tumors by regulating lncRNAs. Lipid metabolites and enzymes involved in lipid metabolism can in turn regulate the expression of lncRNAs to participate in tumor progression. For example, cholesterol regulates lncRNA HULC to promote liver cancer progression, palmitic acid regulates lncRNA UCA1 to promote gastric cancer metastasis, and PLD regulates lncRNA ANRIL to exert an anti-large cell lung cancer effect.
In addition to SphK1, lncRNAs can also regulate Sphk2 to participate in tumorigenesis and development. In papillary thyroid carcinoma (PTC), the expression of LINC00460 is upregulated, which is positively correlated with patients with advanced lymph node metastasis. It was further discovered that LINC00460 can be used as a ceRNA to regulate the expression of SphK2 by sponging miR-613 and promoting cell proliferation, migration and invasion [172]. Another study reported that LINC00520 sponged miR-577 in PTC cells with a similar mechanism to positively regulate Sphk2 expression, leading to increased PTC cell proliferation, migration, invasion and apoptosis [173].
Phospholipase
Phospholipases are enzymes that are involved in the hydrolysis of acyl and phosphate esters of phospholipids. They are important mediators of intracellular and intercellular signaling. Lipid mediators generated by phospholipases regulate multiple cellular processes that can promote cancer development [174].
Phospholipase D (PLD) hydrolyzes phosphatidylcholine (PC) to PA, a signaling lipid, which regulates cell growth and cancer progression through effects on mTOR and PKB/Akt [175]. Thus, PLD expression is increased in cancers, where it correlates with tumor size and prognosis [176, 177]. LINC00511 is upregulated in cervical cancers and could promote PLD1 expression by enriching RXRA to the promoter region of PLD1 and activating PLD1 promoter activity, thus enhancing proliferation and inhibiting the autophagy and apoptosis of cervical cancer cells [178].
Phospholipase A2 (PLA2) catalyzes the hydrolysis of phosphatidic acid, resulting in the production of FFAs and lysophospholipid 1 [179]. There are dozens of secreted PLA2 forms called secreted phospholipase A2 (sPLA2). sPLA2 has oncogenic functions in cancer biology and is usually upregulated in certain types of cancers, such as triple-negative BC [180], NSCLC [181] and ovarian cancer [182]. Studies have found that lncRNA SLNCR1 is highly expressed in NSCLC and may regulate the migration, invasion, and dryness of NSCLC by interacting with sPLA2 [181]. However, further research is needed to determine the mechanisms involved in this process.
Lipid metabolism influences lncRNA expression
The products of lipid metabolism are necessary to support cell survival and development. In the above, lncRNAs affect the expression of metabolites by regulating various enzymes involved in cholesterol metabolism, FA metabolism, TG metabolism, and PL metabolism, thereby affecting the development of cancer. However, lipid metabolites and enzymes involved in lipid metabolism also have a regulatory effect on the expression of lncRNAs, which affects the occurrence and development of tumors (Figure 2 and Table 1).
For example, Cui et al. showed that lncRNA HULC expression was upregulated by cholesterol through a positive feedback loop involving the retinoid receptor RXRA, which activated the HULC promoter [132]. HULC exerts its effect by upregulating the expression of methylase DNA (cytosin-5-)-methyltransferase 1 (DNMT1) and thereby inhibiting the expression of miR-9 by eliciting methylation of CpG islands in the miR-9 promoter, preventing miR-9 from targeting the 3'UTR of PPARA. As a consequence, the transcription level of PPARA in hepatoma cells was upregulated. Given that ACSL1 is activated by the transcription factor PPARA [183], overexpression of ACSL1 further increases the uptake of FAs in HCC cells [184], and its product cholesterol upregulates HULC by activating RXRA. Hence, forming a positive feedback loop to upregulate the level of HULC in hepatoma cells leads to abnormal lipid metabolism and accelerates the occurrence of HCC [132]. LXRs are cholesterol sensors [51]. He et al. found that LXRα can bind to the lncRNA HULC promoter region and inhibit the transcriptional activity of the HULC gene promoter. HULC can inhibit miR-134-5p targeting the 3'-UTR of FOXM1 to upregulate the expression of FOXM1 [185]. In previous studies, FOXM1 has been proven to play a promoting role in a variety of tumors, including HCC [186-188]. Therefore, LXR can inhibit the growth of HCC cells by reducing HULC, increasing miR-134-5p and reducing FOXM1 [185].
Palmitic acid is the most common saturated FA and can be synthesized endogenously de novo or ingested exogenously. It maintains a certain concentration under physiological conditions, maintains membrane stability and participates in protein transport [189]. It has been reported that palmitic acid can promote the nuclear transport of FABP5, thereby increasing the nucleoprotein level of SP1 [190]. SP1 can bind to the promoter of lncRNA UCA1 to increase the expression of UCA1 [191]. Then, UCA1 can regulate GRK2 protein stability by promoting the Cbl-c-mediated ubiquitination and degradation of GRK2, thereby activating the ERK-MMP9 pathway to increase GC cell metastasis [192]. In addition, PLD is an important enzyme involved in regulating phospholipid metabolism. It has been reported that in large cell lung cancer H460 cells, inhibiting PLD expression can stimulate the upregulation of lncRNA ANRIL, promote cell apoptosis, and produce antitumor effects [193].
In addition, Li et al. found that endogenously synthesized n-3 polyunsaturated FAs in fat-1 mice decrease the mammary cancer risk of female offspring by regulating the expression of lncRNAs (53 upregulated and 45 downregulated) [194].
LncRNAs act as regulators of ferroptosis
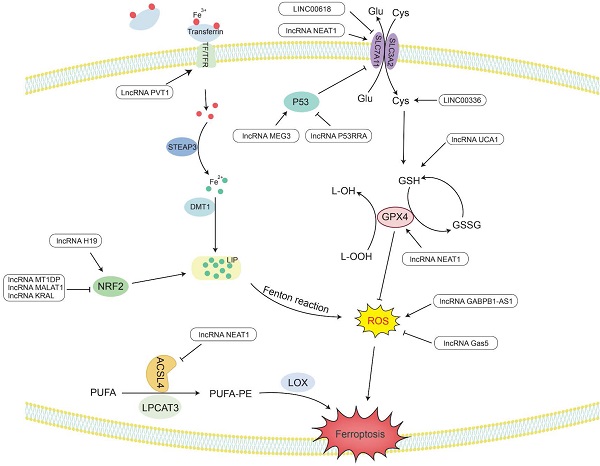
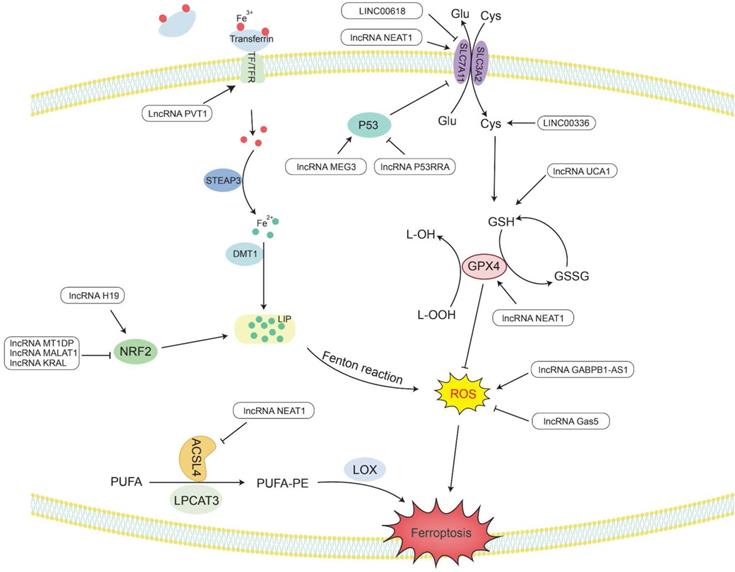
Ferroptosis is a novel type of special cell programmed death that is mainly caused by lipid peroxidation induced by ROS, resulting in lipid peroxide accumulation [8]. In particular, the peroxidation of PUFAs can lead to the formation of lipid hydrogen peroxide (LOOH), which is cleaved under the action of active Fe3+ to form lipid free radicals that are highly reactive and toxic to membranes and cells. These lipid free radicals can extract electrons from adjacent PUFAs, thereby continuing to trigger new LPO and oxidative damage [9]. The inactivation of GPX4 depends on glutathione (GSH), lipid peroxide cannot be catalyzed and reduced by GPX4, and Fe2+-oxidized lipids produce a large amount of ROS by the Fenton reaction, all of which promote ferroptosis [195]. Therefore, studying the mechanism by which lncRNAs affect ferroptosis would provide new ideas for the treatment of cancer (Figure 3 and Table 2).
LncRNAs regulate the activity of GPX4 in ferroptosis
GPX4 is a GSH-dependent enzyme [196], and its antioxidant function requires the use of GSH as a substrate [197], thereby reducing L-OOH to the corresponding nontoxic alcohol, losing its peroxide activity [195], hence reducing ROS and subsequently preventing the occurrence of ferroptosis.
LncRNAs play an indispensable role in regulating ferroptosis. LncRNA UCA1 sponges miR-16 to upregulate the expression of glutaminase 2 (GLS2) in bladder cancer [198]. GLS2 can catalyze the conversion of glutamine into glutamic acid and further synthesize GSH. When glutamine is metabolized, it enters the tricarboxylic acid cycle to produce a large amount of nicotinamide adenine dinucleotide phosphate (NADPH) [199]. This NADPH is required when glutathione reductase (GR) reduces oxidized glutathione (GSSG) to GSH [200]. Therefore, UCA1 can upregulate the expression of GSH and NADPH, making GPX4 play an antioxidant role to reduce the production of ROS and inhibit ferroptosis.
It has been reported that in the human ACSL family, only ACSL4 is related to ferroptosis [201]. Saturated membrane lipids are not sensitive to peroxidation, but ACSL4 can enrich cell membranes with polyunsaturated omega-6 FAs [201, 202]. Therefore, the upregulation of ACSL4 promotes the occurrence of LPO and the sensitivity of cells to ferroptosis [203]. For example, lncRNA NEAT1 can directly bind to ACSL4. Overexpression of NEAT1 can inhibit the expression of ACSL4, increase the expression of SLC7A11 and GPX4, and reduce the content of regulatory iron in cells. Finally, the occurrence of ferroptosis is decreased, and cell apoptosis is inhibited [204].
In addition, nuclear factor erythroid 2-related factor 2 (Nrf2) acts as an antioxidant transcription factor and activates the expression of downstream antioxidant factors. Nrf2 can upregulate genes in the pentose phosphate pathway to produce NADPH, which plays an antioxidant role. Nrf2 can increase the transcription of genes involved in the synthesis of GSH and GPX family genes and increase the activity of GPX4 in ferroptosis to exert antioxidant effects [205]. In ovarian cancer, lncRNA H19 can target Nrf2 to regulate genes related to GSH synthesis. The increase in GSH synthesis enhances the antioxidant effect and promotes cisplatin resistance [206]. In addition, Keap1 (Kelch-like ch-associated protein 1) is a protein that can target the ubiquitination of Nrf1 and accelerate the degradation of Nrf1 in the proteasome [207]. Therefore, there are still some lncRNAs that affect the generation of ROS by regulating the Keap1/Nrf1 pathway. For example, lncRNA KRAL can act as a ceRNA of miR-141 in HCC to upregulate the expression of Keap1, thereby downregulating the expression of Nrf1, weakening the expression of downstream genes in the Nrf2 pathway, and increasing the sensitivity of cancer cells to 5-fluorouracil [207]. Overexpression of lncRNA MALAT1 in multiple myeloma can upregulate Keap1 expression to target Nrf1, causing it to undergo proteasome degradation, leading to the occurrence of oxidative stress [208].
The metabolic pathway of lncRNAs regulating ferroptosis. Lipid peroxides play a key role in initiating ferroptosis. LncRNAs participate in the occurrence of ferroptosis in tumors by regulating related genes involved in lipid peroxide accumulation.
Summary of ferroptosis-associated lncRNAs in cancer
| LncRNA | Associated type of effect | Target | Influence to target | Cancer type | Reference |
|---|---|---|---|---|---|
| LncRNA UCA1 | Ferroptosis | GSH | Up | Bladder cancer | [198] |
| LncRNA NEAT1 | Ferroptosis | ACSL4 | Down | Non-small cell lung cancer | [204] |
| LncRNA NEAT1 | Ferroptosis | SLC7A11/GPX4 | Up | Non-small cell lung cancer | [204] |
| LncRNA H19 | Ferroptosis | Nrf2 | Up | Ovarian cancer | [206] |
| LncRNA KRAL | Ferroptosis | Nrf2 | Down | Hepatocellular carcinoma | [207] |
| LncRNA MALAT1 | Ferroptosis | Nrf2 | Down | Multiple myeloma | [208] |
| LncRNA PVT1 | Ferroptosis | TFR1 | Up | Hepatocellular carcinoma | [212] |
| LncRNA MT1DP | Ferroptosis | Nrf2 | Down | Non-small cell lung cancer | [214] |
| LncRNA P53RRA | Ferroptosis | p53 | Down | Lung cancer | [216] |
| LncRNA MEG3 | Ferroptosis | p53 | Up | Non-small cell lung cancer | [218, 219] |
| LINC00618 | Ferroptosis | SLC7A11 | Down | Acute myeloid leukemia | [220] |
| LINC00336 | Ferroptosis | Cys | Up | Lung cancer | [221] |
| LncRNA GABPB1-AS1 | Ferroptosis | ROS | Up | Hepatocellular carcinoma | [224] |
| LncRNA Gas5 | Ferroptosis | ROS | Down | Melanoma | [225] |
LncRNAs regulate Fe2+ levels in ferroptosis
Maintaining a balance of intracellular Fe levels is essential for cell survival. Fe3+ needs to be taken up into cells under the combined action of transferrin (TF) and transferrin receptor (TFR) [199]. Then, Fe3+ is oxidized to Fe2+ under the redox effect of the six-transmembrane epithelial antigen of prostate 3 (STEAP3) and is ultimately stored in the labile iron pool (LIP) with redox activity and ferritin [209]. In ferroptosis, Fe2+ can provide electrons to oxygen through the Fenton reaction to form ROS [210], which catalyzes the formation of lipid radicals that are highly reactive with biomolecules [195, 211].
LncRNA PVT1 is upregulated in HCC and acts as a sponge for miR-150 to regulate the expression of HIG2, the downstream target gene of miR-150 [212]. HIG2, as a hypoxia-inducing gene, has been determined to be involved in ferroptosis [213]. The detection of iron homeostasis-related protein expression and Fe uptake showed that knocking down PVT1 significantly reduced the expression of TFR1 and increased the expression of ferritin light chain (FTL), ultimately reducing the uptake of Fe and regulating ferroptosis [212].
In addition, Nrf2 can increase the transcription level of ferroportin 1 (FPN1) to increase the export of intracellular iron to the outside of the cell and reduce the level of Fe2+ in the intracellular LIP. Nrf2 can also induce the upregulation of ferritin gene transcription and store excess iron in cells [205]. In NSCLC cells, lncRNA MT1DP inhibits the expression of Nrf2 by stabilizing miR-365a-3p, increases intracellular iron levels, and increases oxidative stress-induced hypertrophy caused by ROS [214].
LncRNAs regulate cysteine metabolism in ferroptosis
SLC7A11 is a plasma membrane transporter that can take extracellular cysteine into cells and transfer intracellular glutamate to the outside of the cell. The increase in intracellular cysteine content increases the synthesis of GSH and promotes GPX4 to exert its antioxidant effect [215].
LncRNA P53RRA has been shown to be mainly located in the cytoplasm and is silent in a variety of cancers, such as NSCLC, liver cancer, colon cancer, NPC and BC. P53RRA can bind to Ras-GTPase activator protein binding protein 1 (G3BP1), resulting in the removal of P53 from the G3BP1 complex and increasing the retention of P53 in the nucleus [216]. P53 inhibits the transcription of SLC7A11 in the nucleus and reduces the uptake of cysteine, consequently limiting the production of GSH in the cell. GSH acts as a cellular antioxidant, and its reduction renders GPX4 unable to function, which can easily lead to the occurrence of LPO, thus increasing the sensitivity of cells to ferroptosis [217]. LncRNA MEG3 was found to be underexpressed in NSCLC, and its overexpression can activate the expression of P53 [218]. Therefore, knocking down MEG3 can reduce P53 and promote the synthesis of GSH, resulting in a decrease in ROS levels and reduced apoptosis [219]. Another study found that LINC00618 can attenuate the expression of lymphoid-specific helicase (LSH) by interacting with LSH and then affect the binding of LSH to the SLC7A11 promoter to reduce the transcriptional activity of SLC7A11 to induce ferroptosis [220]. In addition, LSH can also promote the expression of ELAV-like RNA-binding protein 1 (ELAVL1) by reducing the recruitment of P53 to the ELAVL1 promoter. ELAVL1 can increase its expression in lung cancer by stabilizing the posttranscriptional level of LINC00336. LINC00336 can increase the expression of cystathionine β-synthase (CBS) through the endogenous sponge of miR-6852 [221]. Then, CBS can enhance the transsulfuration pathway (TSP), which converts methionine to cysteine [222]. Finally, glutathione is synthesized to further exert its antioxidant effect [223]. Therefore, LINC00336 can reduce lipid ROS and inhibit the occurrence of ferroptosis through the miR-6852/CBS/GSH pathway [221].
LncRNAs regulate ROS levels through other mechanisms in ferroptosis
In addition to the above factors, lncRNAs also regulate ROS through other mechanisms. High expression of GABPB1 is associated with a poor prognosis of HCC. Erastin is an inducer of ferroptosis that can upregulate the expression of the antisense lncRNA GABPB1-AS1 of the GABPB1 gene, block the transcription and translation level of GABPB1, and inhibit the expression of peroxidase-5 (PRDX5). Therefore, the inhibitory effect of PRDX5 on the production of ROS by H2O2 is weakened, which ultimately inhibits the antioxidant capacity of cancer cells and increases cell death [224]. LncRNA Gas5 was found to induce cell cycle arrest in melanoma. Knockdown of Gas5 can increase the expression of glucose 6-phosphate dehydrogenase (G6PD) and NADPH oxidase 4 (NOX4) [225]. G6PD can promote the production of NADPH, making it function as a substrate for activating NOX4 [226]. NOX4 is a transmembrane protein that produces peroxide and/or hydrogen peroxide, and its upregulation can promote the production of ROS. Therefore, after knocking down Gas5, ferroptosis can occur under the synergistic effect of G6PD and NOX4 [225].
Collectively, these results show that lncRNAs are important regulators of ferroptosis and lipid metabolism.
Conclusion and prospects
A number of studies have revealed that reprogramming and ferroptosis of lipid metabolism are closely related to the occurrence, development and progression of tumors, expanding our understanding of the impact of lipids on tumor etiology.
Today, traditional treatment methods are gradually showing shortcomings, such as nonspecificity and drug resistance. Therefore, the role of lncRNAs in cancer has attracted increasing attention from researchers. In this review, we emphasized lncRNAs related to lipid metabolism, ferroptosis and cancer. Although there have been few studies on the role of lncRNAs in cancer, we can also see promising effects in the treatment of cancer from the limited existing literature. LncRNAs regulate multiple aspects of abnormal tumor lipid metabolism, thus playing a carcinogenic role in multiple aspects of tumorigenesis and affecting tumor cell proliferation, apoptosis, migration, invasion, ferroptosis and other biological processes. LncRNAs can also influence the onset of ferroptosis by regulating related genes in the ferroptosis pathway, enabling them to produce a way to kill cells that is not dependent on apoptosis. Therefore, we believe that targeting lncRNAs may serve as a viable strategy for cancer treatment, creating new opportunities for diagnostic therapeutic interventions in cancer.
However, there are still several problems that remain to be addressed. First, it is still challenging to identify the most important lncRNAs associated with specific types of tumors. Additionally, compared with protein-coding genes, the conservation of lncRNAs across different species is generally poor. Therefore, therapeutic strategies developed using in vitro and animal models cannot be easily extended to human applications immediately and may require further studies.
Taken together, future studies should focus on uncovering the relationships of each lncRNA in tumor lipid metabolism and ferroptosis and developing appropriate model systems to aid in the development of diagnosis, treatment and prognosis in cancer. We believe that with continuous research work, in the future, we could make full use of the advantages of lncRNAs as regulators of tumor metabolism. Great progress will be made in cancer treatment strategies based on lncRNAs to improve the quality of life of many cancer patients.
Abbreviations
FAs: fatty acids; TG: triacylglycerol; PL: phospholipids; LPO: lipid peroxidation; ROS: reactive oxygen species; PUFA: polyunsaturated free fatty acids; lncRNAs: long noncoding RNAs; MVA: mevalonate; HMGCR: hydroxymethylglutaryl-coenzyme A reductase; SQLE: squalene epoxidase; LDLR: low-density lipoprotein receptor; GPX4: glutathione peroxidase 4; SREBP2: sterol regulatory element-binding protein 2; ER: endoplasmic reticulum; BC: breast cancer; BCSC: breast cancer stem cells; PCBP2: poly(rC) binding protein 2; NSCLC: non-small cell lung cancer; CE: cholesterol ester; ACAT: cholesterol acyltransferase; ABC: ATP binding cassette; LXRs: liver X receptors; RXRα: retinoid X receptors alpha; DR: direct repeat; GBM: glioblastoma; ABCA1: ATP-binding cassette transporter A1; MPT: mitochondrial permeability transition; EZH2: enhancer of zeste homolog 2; ACLY: ATP citrate lyase; ACC: acetyl-CoA carboxylase; FASN: fatty acid synthase; SCD: stearoyl-CoA desaturase; CoA: coenzyme A; GC: gastric cancer; NPC: nasopharyngeal carcinoma; SRPK2: serine/arginine protein-specific kinase 2; SR: serine/arginine; MMP-9: matrix metalloproteinase-9; ceRNA: competitive endogenous RNA; FAT/CD36: fatty acid translocase; FATP: fatty acid-binding protein; oxLDL: oxidized low-density lipoprotein; CPT1: carnitine palmityl transferase 1; FAO: fatty acid oxidation; TAM: tumor-associated macrophages; FFA: free fatty acid; HCC: hepatocellular carcinoma; EMT: epithelial-mesenchymal transition; AML: acute myeloid leukemia; TC: thyroid cancer; ACSL: acyl-CoA synthetase long-chain family members; MSCs: mesenchymal stem cells; LD: lipid droplets; LPA: lysophosphatidic acid; GPAT: glycerol 3-phosphate acyltransferase; FA-CoA: fatty acyl-CoA; PA: phosphatidic acid; AGPAT: acylglycerophosphate acyltransferase; PAP or Lipin: phosphatidic acid phosphohydrolase; DG: diacylglycerol; DGAT: diacylglycerol acyltransferase; ATGL: triglyceride lipase; HSL: hormone-sensitive lipase; MAGL: monoacylglycerol lipase; SphKs: sphingosine kinases; S1P: sphingosine-1-phosphate; PTC: papillary thyroid carcinoma; PLD: phospholipase D; PC: phosphatidylcholine; PLA2: phospholipase A2; DNMT1: DNA (Cytosin-5-)-methyltransferase 1; LOOH: lipid hydrogen peroxide; GSH: glutathione; GLS2: glutaminase2; NADPH: nicotinamide adenine dinucleotide phosphate; GR: glutathione reductase; GSSG: oxidized glutathione; Nrf2: Erythroid 2-related factor 2; TF: transferrin; TFR: transferrin receptor; LIP: labile iron pool; STEAP3: six-transmembrane epithelial antigen of the prostate 3; FTL: ferritin light chain; FPN1: ferroportin 1; LSH: lymphoid specific helicase; CBS: cystathionine β-synthase; TSP: transsulfuration pathway; G6PD: glucose 6-phosphate dehydrogenase; NOX4: NADPH oxidase 4.
Acknowledgements
We thank Prof. Yongguang Tao and Dr. Li Xie and Wenbin Liu for verification of the figures and the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81802785 [Y.J.], 82100490[L.C.]), the Hunan Provincial Natural Science Foundation of China (2020JJ5382 [Y.J.], 2020JJ5381 [L.C.]), the Scientific Research Project of Hunan Provincial Health Commission (20200763 [W.L.]) and the Basic Research Project of the Changsha Science and Technology Bureau (kq2004127 [L.X.]).
Author Contributions
L.C. and Y.J. designed/planned and wrote the manuscript. J.H., J.W. and H.H. contributed equally in writing the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Abramson HN. The lipogenesis pathway as a cancer target. J Med Chem. 2011;54(16):5615-5638
2. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610-2623
3. Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond). 2018;38(1):27
4. Pomorski T, Hrafnsdottir S, Devaux PF, van Meer G. Lipid distribution and transport across cellular membranes. Semin Cell Dev Biol. 2001;12(2):139-148
5. Maan M, Peters JM, Dutta M, Patterson AD. Lipid metabolism and lipophagy in cancer. Biochem Biophys Res Commun. 2018;504(3):582-589
6. Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732-749
7. Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis. A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171(2):273-285
8. Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, Kang R, Tang D. Ferroptosis. process and function. Cell Death Differ. 2016;23(3):369-379
9. Cheng Z, Li Y. What is responsible for the initiating chemistry of iron-mediated lipid peroxidation. an update. Chem Rev. 2007;107(3):748-766
10. Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniels VW, Machiels J, Vanderhoydonc F, Smans K, Waelkens E, Verhoeven G, Swinnen JV. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Research. 2010;70(20):8117-8126
11. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253-1261
12. Alessio E, Bonadio RS, Buson L, Chemello F, Cagnin S. A Single Cell but Many Different Transcripts. A Journey into the World of Long Non-Coding RNAs. Int J Mol Sci. 2020 21(1)
13. Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11(3):151-160
14. Vendramin R, Marine JC, Leucci E. Non-coding RNAs. the dark side of nuclear-mitochondrial communication. EMBO J. 2017;36(9):1123-1133
15. Jie Y, Ye L, Chen H, Yu X, Cai L, He W, Fu Y. ELFN1-AS1 accelerates cell proliferation, invasion and migration via regulating miR-497-3p/CLDN4 axis in ovarian cancer. Bioengineered. 2020;11(1):872-882
16. Wang Y, Zhou P, Li P, Yang F, Gao XQ. Long non-coding RNA H19 regulates proliferation and doxorubicin resistance in MCF-7 cells by targeting PARP1. Bioengineered. 2020;11(1):536-546
17. Ponzio G, Rezzonico R, Bourget I, Allan R, Nottet N, Popa A, Magnone V, Rios G, Mari B, Barbry P. A new long noncoding RNA (lncRNA) is induced in cutaneous squamous cell carcinoma and down-regulates several anticancer and cell differentiation genes in mouse. J Biol Chem. 2017;292(30):12483-12495
18. Gao B, Li S, Li G. Long Noncoding RNA (lncRNA) Small Nucleolar RNA Host Gene 5 (SNHG5) Regulates Proliferation, Differentiation, and Apoptosis of K562 Cells in Chronic Myeliod Leukemia. Med Sci Monit. 2019;25:6812-6819
19. Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29(6):e95
20. Dasgupta P, Kulkarni P, Majid S, Hashimoto Y, Shiina M, Shahryari V, Bhat NS, Tabatabai L, Yamamura S, Saini S, Tanaka Y, Dahiya R. LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis. 2020;11(8):660
21. Ren Z, Liu X, Si Y, Yang D. Long non-coding RNA DDX11-AS1 facilitates gastric cancer progression by regulating miR-873-5p/SPC18 axis. Artif Cells Nanomed Biotechnol. 2020;48(1):572-583
22. Fan Y, Sheng W, Meng Y, Cao Y, Li R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif Cells Nanomed Biotechnol. 2020;48(1):393-407
23. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21(4):225-245
24. Riscal R, Skuli N, Simon MC. Even Cancer Cells Watch Their Cholesterol!. Mol Cell. 2019;76(2):220-231
25. Ding X, Zhang W, Li S, Yang H. The role of cholesterol metabolism in cancer. Am J Cancer Res. 2019;9(2):219-227
26. Huang B, Song BL, Xu C. Cholesterol metabolism in cancer. mechanisms and therapeutic opportunities. Nat Metab. 2020;2(2):132-141
27. Sun H, Huang Z, Sheng W, Xu MD. Emerging roles of long non-coding RNAs in tumor metabolism. J Hematol Oncol. 2018;11(1):106
28. Meaney S. Epigenetic regulation of cholesterol homeostasis. Front Genet. 2014;5:311
29. Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys. 2011;505(2):131-143
30. Do R, Kiss RS, Gaudet D, Engert JC. Squalene synthase. a critical enzyme in the cholesterol biosynthesis pathway. Clin Genet. 2009;75(1):19-29
31. Gobel A, Rauner M, Hofbauer LC, Rachner TD. Cholesterol and beyond - The role of the mevalonate pathway in cancer biology. Biochim Biophys Acta Rev Cancer. 2020;1873(2):188351
32. Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34-47
33. Bathaie SZ, Ashrafi M, Azizian M, Tamanoi F. Mevalonate Pathway and Human Cancers. Curr Mol Pharmacol. 2017;10(2):77-85
34. Waller DD, Park J, Tsantrizos YS. Inhibition of farnesyl pyrophosphate (FPP) and/or geranylgeranyl pyrophosphate (GGPP) biosynthesis and its implication in the treatment of cancers. Crit Rev Biochem Mol Biol. 2019;54(1):41-60
35. Tetali SD. Terpenes and isoprenoids. a wealth of compounds for global use. Planta. 2019;249(1):1-8
36. Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425-430
37. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism. New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188394
38. Zielinski ZA, Pratt DA. Cholesterol Autoxidation Revisited. Debunking the Dogma Associated with the Most Vilified of Lipids. J Am Chem Soc. 2016;138(22):6932-6935
39. Forcina GC, Dixon SJ. GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis. Proteomics. 2019;19(18):e1800311
40. Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9(2):125-138
41. Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors. master regulators of lipid homeostasis. Biochimie. 2004;86(11):839-848
42. Horton JD, Goldstein JL, Brown MS. SREBPs. activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125-1131
43. Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 2004;23(4):950-958
44. Gabitova L, Gorin A, Astsaturov I. Molecular pathways. sterols and receptor signaling in cancer. Clin Cancer Res. 2014;20(1):28-34
45. Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH, Yu Y. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15(33):3831-3844
46. Brown DN, Caffa I, Cirmena G, Piras D, Garuti A, Gallo M, Alberti S, Nencioni A, Ballestrero A, Zoppoli G. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep. 2016;6:19435
47. Qin Y, Hou Y, Liu S, Zhu P, Wan X, Zhao M, Peng M, Zeng H, Li Q, Jin T, Cui X, Liu M. A Novel Long Non-Coding RNA lnc030 Maintains Breast Cancer Stem Cell Stemness by Stabilizing SQLE mRNA and Increasing Cholesterol Synthesis. Adv Sci (Weinh). 2021;8(2):2002232
48. Wang L, Lin C, Sun N, Wang Q, Ding X, Sun Y. Long non-coding RNA CASC19 facilitates non-small cell lung cancer cell proliferation and metastasis by targeting the miR-301b-3p/LDLR axis. J Gene Med. 2020;22(12):e3254
49. Sharma B, Agnihotri N. Role of cholesterol homeostasis and its efflux pathways in cancer progression. J Steroid Biochem Mol Biol. 2019;191:105377
50. Wang YJ, Bian Y, Luo J, Lu M, Xiong Y, Guo SY, Yin HY, Lin X, Li Q, Chang CCY, Chang TY, Li BL, Song BL. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat Cell Biol. 2017;19(7):808-819
51. van Solingen C, Scacalossi KR, Moore KJ. Long noncoding RNAs in lipid metabolism. Curr Opin Lipidol. 2018;29(3):224-232
52. Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116(3):607-614
53. Edwards PA, Kast HR, Anisfeld AM. BAREing it all. the adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43(1):2-12
54. Schmitz G, Langmann T. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochim Biophys Acta. 2005;1735(1):1-19
55. Vedin LL, Lewandowski SA, Parini P, Gustafsson JA, Steffensen KR. The oxysterol receptor LXR inhibits proliferation of human breast cancer cells. Carcinogenesis. 2009;30(4):575-579
56. Lo Sasso G, Bovenga F, Murzilli S, Salvatore L, Di Tullio G, Martelli N, D'Orazio A, Rainaldi S, Vacca M, Mangia A, Palasciano G, Moschetta A. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology. 2013;144(7):1497-1507 1507 e1491-1413
57. Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, Rong X, Hong C, Turner KM, Liu F, Hon GC, Jenkins D, Martini M, Armando AM, Quehenberger O, Cloughesy TF, Furnari FB, Cavenee WK, Tontonoz P, Gahman TC, Shiau AK, Cravatt BF, Mischel PS. An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers. Cancer Cell. 2016;30(5):683-693
58. Lee SD, Tontonoz P. Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis. 2015;242(1):29-36
59. Montero J, Morales A, Llacuna L, Lluis JM, Terrones O, Basanez G, Antonsson B, Prieto J, Garcia-Ruiz C, Colell A, Fernandez-Checa JC. Mitochondrial cholesterol contributes to chemotherapy resistance in hepatocellular carcinoma. Cancer Research. 2008;68(13):5246-5256
60. Meng XD, Yao HH, Wang LM, Yu M, Shi S, Yuan ZX, Liu J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE(-/-) Mice. Mol Ther Nucleic Acids. 2020;19:84-96
61. Cai C, Zhu H, Ning X, Li L, Yang B, Chen S, Wang L, Lu X, Gu D. LncRNA ENST00000602558.1 regulates ABCG1 expression and cholesterol efflux from vascular smooth muscle cells through a p65-dependent pathway. Atherosclerosis. 2019;285:31-39
62. Chirala SS, Chang H, Matzuk M, Abu-Elheiga L, Mao J, Mahon K, Finegold M, Wakil SJ. Fatty acid synthesis is essential in embryonic development. fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci U S A. 2003;100(11):6358-6363
63. Cao Y, Wang S, Liu S, Wang Y, Jin H, Ma H, Luo X, Cao Y, Lian Z. Effects of Long-Chain Fatty Acyl-CoA Synthetase 1 on Diglyceride Synthesis and Arachidonic Acid Metabolism in Sheep Adipocytes. Int J Mol Sci. 2020 21(6)
64. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18(2):153-161
65. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism. fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227-232
66. Shearn CT, Mercer KE, Orlicky DJ, Hennings L, Smathers-McCullough RL, Stiles BL, Ronis MJ, Petersen DR. Short term feeding of a high fat diet exerts an additive effect on hepatocellular damage and steatosis in liver-specific PTEN knockout mice. PLoS One. 2014;9(5):e96553
67. Lupu R, Menendez JA. Targeting fatty acid synthase in breast and endometrial cancer. An alternative to selective estrogen receptor modulators? Endocrinology. 2006;147(9):4056-4066
68. Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6(4):551-562
69. Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol. 1984;247(1 Pt 2):R146-153
70. Chen M, Huang J. The expanded role of fatty acid metabolism in cancer. new aspects and targets. Precis Clin Med. 2019;2(3):183-191
71. Khwairakpam AD, Shyamananda MS, Sailo BL, Rathnakaram SR, Padmavathi G, Kotoky J, Kunnumakkara AB. ATP citrate lyase (ACLY). a promising target for cancer prevention and treatment. Curr Drug Targets. 2015;16(2):156-163
72. Granchi C. ATP citrate lyase (ACLY) inhibitors. An anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur J Med Chem. 2018;157:1276-1291
73. Teng L, Chen Y, Cao Y, Wang W, Xu Y, Wang Y, Lv J, Li C, Su Y. Overexpression of ATP citrate lyase in renal cell carcinoma tissues and its effect on the human renal carcinoma cells in vitro. Oncol Lett. 2018;15(5):6967-6974
74. Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, Sidoli S, Parris JLD, Affronti HC, Sivanand S, Egolf S, Sela Y, Trizzino M, Gardini A, Garcia BA, Snyder NW, Stanger BZ, Wellen KE. Acetyl-CoA Metabolism Supports Multistep Pancreatic Tumorigenesis. Cancer Discov. 2019;9(3):416-435
75. Koufaris C, Valbuena GN, Pomyen Y, Tredwell GD, Nevedomskaya E, Lau CH, Yang T, Benito A, Ellis JK, Keun HC. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene. 2016;35(21):2766-2776
76. Cheng Y, Jia B, Wang Y, Wan S. miR-133b acts as a tumor suppressor and negatively regulates ATP citrate lyase via PPARgamma in gastric cancer. Oncol Rep. 2017;38(5):3220-3226
77. Zheng ZQ, Li ZX, Guan JL, Liu X, Li JY, Chen Y, Lin L, Kou J, Lv JW, Zhang LL, Zhou GQ, Liu RQ, Chen F, He XJ, Li YQ, Li F, Xu SS, Ma J, Liu N, Sun Y. Long Noncoding RNA TINCR-Mediated Regulation of Acetyl-CoA Metabolism Promotes Nasopharyngeal Carcinoma Progression and Chemoresistance. Cancer Research. 2020;80(23):5174-5188
78. Zhang G, Wang Q, Lu J, Ma G, Ge Y, Chu H, Du M, Wang M, Zhang Z. Long non-coding RNA FLJ22763 is involved in the progression and prognosis of gastric cancer. Gene. 2019;693:84-91
79. Zheng X, Wang X, Zheng L, Zhao H, Li W, Wang B, Xue L, Tian Y, Xie Y. Construction and Analysis of the Tumor-Specific mRNA-miRNA-lncRNA Network in Gastric Cancer. Front Pharmacol. 2020;11:1112
80. Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A. 2005;102(7):2561-2566
81. Khatun M, Sur S, Steele R, Ray R, Ray RB. Inhibition of Long Noncoding RNA Linc-Pint by Hepatitis C Virus in Infected Hepatocytes Enhances Lipogenesis. Hepatology. 2020
82. Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122(3):191-207
83. Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, Chavez A, Zhang E, Isik M, Couvillon A, Dephoure NE, Blackwell TK, Yu JJ, Rabinowitz JD, Cantley LC, Blenis J. Post-transcriptional Regulation of De novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell. 2017;171(7):1545-1558 e1518
84. Cruz-Gil S, Fernandez LP, Sanchez-Martinez R, Gomez de Cedron M, Ramirez de Molina A. Non-Coding and Regulatory RNAs as Epigenetic Remodelers of Fatty Acid Homeostasis in Cancer. Cancers (Basel). 2020 12(10)
85. Wang C, Meng X, Zhou Y, Yu J, Li Q, Liao Z, Gu Y, Han J, Linghu S, Jiao Z, Wang T, Zhang CY, Chen X. Long noncoding RNA CTD-2245E15.3 promotes anabolic enzymes ACC1 and PC to support non-small cell lung cancer growth. Cancer Research 2021
86. Li CJ, Chiu YH, Chang C, Chang YI, Sheu JJ, Chiang AJ. Acetyl Coenzyme A Synthase 2 Acts as a Prognostic Biomarker Associated with Immune Infiltration in Cervical Squamous Cell Carcinoma. Cancers (Basel). 2021 13(13)
87. Tang Y, Tang R, Tang M, Huang P, Liao Z, Zhou J, Zhou L, Su M, Chen P, Jiang J, Hu Y, Zhou Y, Liao Q, Zeng Z, Xiong W, Chen J, Nie S. LncRNA DNAJC3-AS1 Regulates Fatty Acid Synthase via the EGFR Pathway to Promote the Progression of Colorectal Cancer. Front Oncol. 2020;10:604534
88. Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311(5765):1258-1262
89. Menendez JA, Lupu R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin Ther Targets. 2017;21(11):1001-1016
90. Ali A, Levantini E, Teo JT, Goggi J, Clohessy JG, Wu CS, Chen L, Yang H, Krishnan I, Kocher O, Zhang J, Soo RA, Bhakoo K, Chin TM, Tenen DG. Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non-small cell lung cancer. EMBO Mol Med. 2018 10(3)
91. Sun W, Nie W, Wang Z, Zhang H, Li Y, Fang X. Lnc HAGLR Promotes Colon Cancer Progression Through Sponging miR-185-5p and Activating CDK4 and CDK6 in vitro and in vivo. Onco Targets Ther. 2020;13:5913-5925
92. Li YH, Huang GM, Wang W, Zang HL. LncRNA HAGLR exacerbates hepatocellular carcinoma through negatively regulating miR-6785-5p. Eur Rev Med Pharmacol Sci. 2020;24(18):9353-9360
93. Zhang C, Yang Y, Yi L, Paizula X, Xu W, Wu X. HOXD Antisense Growth-Associated Long Noncoding RNA Promotes Triple-Negative Breast Cancer Progression by Activating Wnt Signaling Pathway. J Breast Cancer. 2021;24(3):315-329
94. Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol. 2017;39(4):1010428317697574
95. Bhan A, Mandal SS. LncRNA HOTAIR. A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151-164
96. Ma DD, Yuan LL, Lin LQ. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci. 2017;21(22):5143-5152
97. Huang H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors. Recent Advances. Sensors (Basel). 2018 18(10)
98. Zhou Q, Chen F, Zhao J, Li B, Liang Y, Pan W, Zhang S, Wang X, Zheng D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7(50):82620-82633
99. Noto A, Raffa S, De Vitis C, Roscilli G, Malpicci D, Coluccia P, Di Napoli A, Ricci A, Giovagnoli MR, Aurisicchio L, Torrisi MR, Ciliberto G, Mancini R. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947
100. Fritz V, Benfodda Z, Rodier G, Henriquet C, Iborra F, Avances C, Allory Y, de la Taille A, Culine S, Blancou H, Cristol JP, Michel F, Sardet C, Fajas L. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol Cancer Ther. 2010;9(6):1740-1754
101. Du X, Wang QR, Chan E, Merchant M, Liu J, French D, Ashkenazi A, Qing J. FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Research. 2012;72(22):5843-5855
102. Hess D, Igal RA. Genistein downregulates de novo lipid synthesis and impairs cell proliferation in human lung cancer cells. Exp Biol Med (Maywood). 2011;236(6):707-713
103. von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, Tan WW, Tun HW, Copland JA. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19(9):2368-2380
104. Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen JB, Pedersen JS, Lund AH, Vang S, Stribolt K, Madsen MR, Laurberg S, McGuire SE, Orntoft TF, Andersen CL. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10(8):1266-1282
105. Guo J, Fang W, Sun L, Lu Y, Dou L, Huang X, Tang W, Yu L, Li J. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat Commun. 2018;9(1):612
106. Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. 2015;5:17454
107. Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M, Kozuka-Hata H, Hiyoshi M, Kitayama J, Negishi L, Kawasaki Y, Akiyama T. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc Natl Acad Sci U S A. 2016;113(5):1273-1278
108. Wang J, Li Y. CD36 tango in cancer. signaling pathways and functions. Theranostics. 2019;9(17):4893-4908
109. Nicholson AC. Expression of CD36 in macrophages and atherosclerosis. the role of lipid regulation of PPARgamma signaling. Trends Cardiovasc Med. 2004;14(1):8-12
110. Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23-35
111. Geeraerts X, Bolli E, Fendt SM, Van Ginderachter JA. Macrophage Metabolism As Therapeutic Target for Cancer, Atherosclerosis, and Obesity. Front Immunol. 2017;8:289
112. Stoger JL, Gijbels MJ, van der Velden S, Manca M, van der Loos CM, Biessen EA, Daemen MJ, Lutgens E, de Winther MP. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis. 2012;225(2):461-468
113. Ruytinx P, Proost P, Van Damme J, Struyf S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front Immunol. 2018;9:1930
114. Huangfu N, Xu Z, Zheng W, Wang Y, Cheng J, Chen X. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating beta-catenin. Biochem Biophys Res Commun. 2018;495(3):2111-2117
115. Hung J, Scanlon JP, Mahmoud AD, Rodor J, Ballantyne M, Fontaine MAC, Temmerman L, Kaczynski J, Connor KL, Bhushan R, Biessen EAL, Newby DE, Sluimer JC, Baker AH. Novel Plaque Enriched Long Noncoding RNA in Atherosclerotic Macrophage Regulation (PELATON). Arterioscler Thromb Vasc Biol. 2020;40(3):697-713
116. Liu YC, Zou XB, Chai YF, Yao YM. Macrophage polarization in inflammatory diseases. Int J Biol Sci. 2014;10(5):520-529
117. Ohata T, Yokoo H, Kamiyama T, Fukai M, Aiyama T, Hatanaka Y, Hatanaka K, Wakayama K, Orimo T, Kakisaka T, Kobayashi N, Matsuno Y, Taketomi A. Fatty acid-binding protein 5 function in hepatocellular carcinoma through induction of epithelial-mesenchymal transition. Cancer Med. 2017;6(5):1049-1061
118. Levi L, Lobo G, Doud MK, von Lintig J, Seachrist D, Tochtrop GP, Noy N. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Research. 2013;73(15):4770-4780
119. Adamson J, Morgan EA, Beesley C, Mei Y, Foster CS, Fujii H, Rudland PS, Smith PH, Ke Y. High-level expression of cutaneous fatty acid-binding protein in prostatic carcinomas and its effect on tumorigenicity. Oncogene. 2003;22(18):2739-2749
120. Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, Huang J, Liang Y, Liu J, Zhao Y, Guo L, Hu Z, Yao S. LNMICC Promotes Nodal Metastasis of Cervical Cancer by Reprogramming Fatty Acid Metabolism. Cancer Research. 2018;78(4):877-890
121. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, Liu Z, Yao Z, Wu Q, Liao W, Zhang S, Liu Y, Xiang Y, Liu J, Shi M. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38(23):4637-4654
122. Han J, Qu H, Han M, Ding Y, Xie M, Hu J, Chen Y, Dong H. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene. 2021;40(4):833-847
123. Lin H, Patel S, Affleck VS, Wilson I, Turnbull DM, Joshi AR, Maxwell R, Stoll EA. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro Oncol. 2017;19(1):43-54
124. Wu Y, Hurren R, MacLean N, Gronda M, Jitkova Y, Sukhai MA, Minden MD, Schimmer AD. Carnitine transporter CT2 (SLC22A16) is over-expressed in acute myeloid leukemia (AML) and target knockdown reduces growth and viability of AML cells. Apoptosis. 2015;20(8):1099-1108
125. Qu Q, Zeng F, Liu X, Wang QJ, Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I. emerging therapeutic targets in cancer. Cell Death Dis. 2016;7:e2226
126. Tang Y, Zhou J, Hooi SC, Jiang YM, Lu GD. Fatty acid activation in carcinogenesis and cancer development. Essential roles of long-chain acyl-CoA synthetases. Oncol Lett. 2018;16(2):1390-1396
127. Mashek DG, Bornfeldt KE, Coleman RA, Berger J, Bernlohr DA, Black P, DiRusso CC, Farber SA, Guo W, Hashimoto N, Khodiyar V, Kuypers FA, Maltais LJ, Nebert DW, Renieri A, Schaffer JE, Stahl A, Watkins PA, Vasiliou V, Yamamoto TT. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J Lipid Res. 2004;45(10):1958-1961
128. Cao Y, Dave KB, Doan TP, Prescott SM. Fatty acid CoA ligase 4 is up-regulated in colon adenocarcinoma. Cancer Research. 2001;61(23):8429-8434
129. Ye X, Zhang Y, Wang X, Li Y, Gao Y. Tumor-suppressive functions of long-chain acyl-CoA synthetase 4 in gastric cancer. IUBMB Life. 2016;68(4):320-327
130. Guo L, Lu J, Gao J, Li M, Wang H, Zhan X. The function of SNHG7/miR-449a/ACSL1 axis in thyroid cancer. J Cell Biochem. 2020;121(10):4034-4042
131. Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia Y, Xu Y, Ma B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2020;65:109422
132. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Research. 2015;75(5):846-857
133. Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350-1358
134. Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2011;1807(6):726-734
135. Tan Y, Lin K, Zhao Y, Wu Q, Chen D, Wang J, Liang Y, Li J, Hu J, Wang H, Liu Y, Zhang S, He W, Huang Q, Hu X, Yao Z, Liang B, Liao W, Shi M. Adipocytes fuel gastric cancer omental metastasis via PITPNC1-mediated fatty acid metabolic reprogramming. Theranostics. 2018;8(19):5452-5468
136. Xie QP, Xiang C, Wang G, Lei KF, Wang Y. MACC1 upregulation promotes gastric cancer tumor cell metastasis and predicts a poor prognosis. J Zhejiang Univ Sci B. 2016;17(5):361-366
137. Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, Birchmeier W, Schlag PM. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15(1):59-67
138. Wang L, Wu Y, Lin L, Liu P, Huang H, Liao W, Zheng D, Zuo Q, Sun L, Huang N, Shi M, Liao Y, Liao W. Metastasis-associated in colon cancer-1 upregulation predicts a poor prognosis of gastric cancer, and promotes tumor cell proliferation and invasion. Int J Cancer. 2013;133(6):1419-1430
139. Zhang R, Shi H, Chen Z, Wu Q, Ren F, Huang H. Effects of metastasis-associated in colon cancer 1 inhibition by small hairpin RNA on ovarian carcinoma OVCAR-3 cells. J Exp Clin Cancer Res. 2011;30:83
140. Hagemann C, Fuchs S, Monoranu CM, Herrmann P, Smith J, Hohmann T, Grabiec U, Kessler AF, Dehghani F, Lohr M, Ernestus RI, Vince GH, Stein U. Impact of MACC1 on human malignant glioma progression and patients' unfavorable prognosis. Neuro Oncol. 2013;15(12):1696-1709
141. Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29(2):157-168
142. Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11(4):233
143. Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals. a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405-1428
144. Xiong Y, Liu Z, Li Z, Wang S, Shen N, Xin Y, Huang T. Long noncoding RNA nuclear paraspeckle assembly transcript 1 interacts with microRNA107 to modulate breast cancer growth and metastasis by targeting carnitine palmitoyltransferase1. Int J Oncol. 2019;55(5):1125-1136
145. Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism. synthesis, lipolysis, and signaling. Chem Rev. 2011;111(10):6359-6386
146. Petan T, Jarc E, Jusovic M. Lipid Droplets in Cancer. Guardians of Fat in a Stressful World. Molecules. 2018 23(8)
147. Yu XH, Wang HF, Wu JB, Wang SS, Tang YJ, Tang YL, Liang XH. Non-coding RNAs derailed. The many influences on the fatty acid reprogramming of cancer. Life Sci. 2019;231:116509
148. Yamashita A, Hayashi Y, Matsumoto N, Nemoto-Sasaki Y, Oka S, Tanikawa T, Sugiura T. Glycerophosphate/Acylglycerophosphate acyltransferases. Biology (Basel). 2014;3(4):801-830
149. Hapala I, Marza E, Ferreira T. Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol Cell. 2011;103(6):271-285
150. Mazar J, Zhao W, Khalil AM, Lee B, Shelley J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S, Finck BN, Han X, Mattick JS, Dinger ME, Perera RJ. The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget. 2014;5(19):8959-8969
151. Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95(11):5987-5992
152. Li D, Guo L, Deng B, Li M, Yang T, Yang F, Yang Z. Long noncoding RNA HR1 participates in the expression of SREBP1c through phosphorylation of the PDK1/AKT/FoxO1 pathway. Mol Med Rep. 2018;18(3):2850-2856
153. Kamei Y, Miura S, Suganami T, Akaike F, Kanai S, Sugita S, Katsumata A, Aburatani H, Unterman TG, Ezaki O, Ogawa Y. Regulation of SREBP1c gene expression in skeletal muscle. role of retinoid X receptor/liver X receptor and forkhead-O1 transcription factor. Endocrinology. 2008;149(5):2293-2305
154. Cerk IK, Wechselberger L, Oberer M. Adipose Triglyceride Lipase Regulation. An Overview. Curr Protein Pept Sci. 2018;19(2):221-233
155. Vegliante R, Di Leo L, Ciccarone F, Ciriolo MR. Hints on ATGL implications in cancer. beyond bioenergetic clues. Cell Death Dis. 2018;9(3):316
156. Wang YY, Attane C, Milhas D, Dirat B, Dauvillier S, Guerard A, Gilhodes J, Lazar I, Alet N, Laurent V, Le Gonidec S, Biard D, Herve C, Bost F, Ren GS, Bono F, Escourrou G, Prentki M, Nieto L, Valet P, Muller C. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2(4):e87489
157. Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6(29):28282-28295
158. Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, Pan S, Zhu M, Liu Y, Wang Y, Meng F, Cui Y, Wang J, Zhang B, Song X, Lu Z, Zheng T, Liu L. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018;17(1):90
159. Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138(3):476-488
160. Bandu R, Mok HJ, Kim KP. Phospholipids as cancer biomarkers. Mass spectrometry-based analysis. Mass Spectrom Rev. 2018;37(2):107-138
161. Pitson SM, Moretti PA, Zebol JR, Lynn HE, Xia P, Vadas MA, Wattenberg BW. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22(20):5491-5500
162. Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325(5945):1254-1257
163. Strub GM, Paillard M, Liang J, Gomez L, Allegood JC, Hait NC, Maceyka M, Price MM, Chen Q, Simpson DC, Kordula T, Milstien S, Lesnefsky EJ, Spiegel S. Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 2011;25(2):600-612
164. Spiegel S, Milstien S. Sphingosine-1-phosphate. an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4(5):397-407
165. Kawamori T, Kaneshiro T, Okumura M, Maalouf S, Uflacker A, Bielawski J, Hannun YA, Obeid LM. Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J. 2009;23(2):405-414
166. Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, Ogretmen B. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J Biol Chem. 2007;282(15):10922-10934
167. Wang F, Wu Z. Sphingosine kinase 1 overexpression is associated with poor prognosis and oxaliplatin resistance in hepatocellular carcinoma. Exp Ther Med. 2018;15(6):5371-5376
168. Hasanifard L, Sheervalilou R, Majidinia M, Yousefi B. New insights into the roles and regulation of SphK2 as a therapeutic target in cancer chemoresistance. J Cell Physiol. 2019;234(6):8162-8181
169. Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60(4):626-636
170. Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z, Li J, Ye L, Zhang X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget. 2016;7(1):241-254
171. Wang L, Chen X, Sun X, Suo J. Long Noncoding RNA LINC00460 Facilitates Colorectal Cancer Progression by Negatively Regulating miR-613. Onco Targets Ther. 2020;13:7555-7569
172. Feng L, Yang B, Tang XD. Long noncoding RNA LINC00460 promotes carcinogenesis via sponging miR-613 in papillary thyroid carcinoma. J Cell Physiol. 2019;234(7):11431-11439
173. Sun Y, Shi T, Ma Y, Qin H, Li K. Long noncoding RNA LINC00520 accelerates progression of papillary thyroid carcinoma by serving as a competing endogenous RNA of microRNA-577 to increase Sphk2 expression. Cell Cycle. 2020;19(7):787-800
174. Park JB, Lee CS, Jang JH, Ghim J, Kim YJ, You S, Hwang D, Suh PG, Ryu SH. Phospholipase signalling networks in cancer. Nat Rev Cancer. 2012;12(11):782-792
175. Zhou W, Shi K, Ji L, Wu R, Chen Y, Tu H, Zhou B, Wang Z, Zhang M. Inhibition of Phospholipase D1 mRNA Expression Slows Down the Proliferation Rate of Prostate Cancer Cells That Have Transited to Androgen Independence. J Cancer. 2018;9(19):3620-3625
176. Noble AR, Maitland NJ, Berney DM, Rumsby MG. Phospholipase D inhibitors reduce human prostate cancer cell proliferation and colony formation. Br J Cancer. 2018;118(2):189-199
177. Gomez-Cambronero J. Lack of effective translational regulation of PLD expression and exosome biogenesis in triple-negative breast cancer cells. Cancer Metastasis Rev. 2018;37(2-3):491-507
178. Shi Y, Liu M, Huang Y, Zhang J, Yin L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J Cell Physiol. 2020;235(10):6592-6604
179. Sved P, Scott KF, McLeod D, King NJ, Singh J, Tsatralis T, Nikolov B, Boulas J, Nallan L, Gelb MH, Sajinovic M, Graham GG, Russell PJ, Dong Q. Oncogenic action of secreted phospholipase A2 in prostate cancer. Cancer Research. 2004;64(19):6934-6940
180. Najahi-Missaoui W, Quach ND, Somanath PR, Cummings BS. Liposomes Targeting P21 Activated Kinase-1 (PAK-1) and Selective for Secretory Phospholipase A2 (sPLA2) Decrease Cell Viability and Induce Apoptosis in Metastatic Triple-Negative Breast Cancer Cells. Int J Mol Sci. 2020 21(24)
181. Xu W, Xu Q, Kuang D, Wang Z, Lu Q, Lin Q, Wu H, Chen L. Long noncoding RNA SLNCR1 regulates nonsmall cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol Med Rep. 2019;20(3):2591-2596
182. Ray U, Roy D, Jin L, Thirusangu P, Staub J, Xiao Y, Kalogera E, Wahner Hendrickson AE, Cullen GD, Goergen K, Oberg AL, Shridhar V. Group III phospholipase A2 downregulation attenuated survival and metastasis in ovarian cancer and promotes chemo-sensitization. J Exp Clin Cancer Res. 2021;40(1):182
183. Phillips CM, Goumidi L, Bertrais S, Field MR, Cupples LA, Ordovas JM, Defoort C, Lovegrove JA, Drevon CA, Gibney MJ, Blaak EE, Kiec-Wilk B, Karlstrom B, Lopez-Miranda J, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J Lipid Res. 2010;51(7):1793-1800
184. Krammer J, Digel M, Ehehalt F, Stremmel W, Fullekrug J, Ehehalt R. Overexpression of CD36 and acyl-CoA synthetases FATP2, FATP4 and ACSL1 increases fatty acid uptake in human hepatoma cells. Int J Med Sci. 2011;8(7):599-614
185. He J, Yang T, He W, Jiang S, Zhong D, Xu Z, Wei Q, Zhang Y, Shi C. Liver X receptor inhibits the growth of hepatocellular carcinoma cells via regulating HULC/miR-134-5p/FOXM1 axis. Cell Signal. 2020;74:109720
186. Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, Nie YZ, Wu KC, Yang AG, Zhang R. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500(4):924-929
187. Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1. Repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52(Pt 1):74-84
188. Gartel AL. FOXM1 in Cancer. Interactions and Vulnerabilities. Cancer Research. 2017;77(12):3135-3139
189. Carta G, Murru E, Banni S, Manca C. Palmitic Acid. Physiological Role, Metabolism and Nutritional Implications. Front Physiol. 2017;8:902
190. Pan J, Dai Q, Zhang T, Li C. Palmitate acid promotes gastric cancer metastasis via FABP5/SP1/UCA1 pathway. Cancer Cell Int. 2019;19:69
191. Wang ZQ, Cai Q, Hu L, He CY, Li JF, Quan ZW, Liu BY, Li C, Zhu ZG. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis. 2017;8(6):e2839
192. Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu QL, Su LP, Liu BY, Li C, Zhu Z. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10-21
193. Kang YH, Kim D, Jin EJ. Down-regulation of Phospholipase D Stimulates Death of Lung Cancer Cells Involving Up-regulation of the Long ncRNA ANRIL. Anticancer Res. 2015;35(5):2795-2803
194. Li J, Li K, Gao J, Guo X, Lu M, Li Z, Li D. Endogenously Synthesized n-3 Polyunsaturated Fatty Acids in Pregnant fat-1 Mice Decreases Mammary Cancer Risk of Female Offspring by Regulating Expression of Long Noncoding RNAs. Mol Nutr Food Res. 2019;63(6):e1801150
195. Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cellular and Molecular Life Sciences. 2016;73(11-12):2195-2209
196. Hsu JL, Chou JW, Chen TF, Hsu JT, Su FY, Lan JL, Wu PC, Hu CM, Lee EY, Lee WH. Glutathione peroxidase 8 negatively regulates caspase-4/11 to protect against colitis. EMBO Mol Med. 2020;12(1):e9386
197. Friedmann Angeli JP, Conrad M. Selenium and GPX4, a vital symbiosis. Free Radic Biol Med. 2018;127:153-159
198. Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J Clin Oncol. 2015;45(11):1055-1063
199. Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell. 2015;59(2):298-308
200. Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830(5):3217-3266
201. Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, Prokisch H, Trumbach D, Mao G, Qu F, Bayir H, Fullekrug J, Scheel CH, Wurst W, Schick JA, Kagan VE, Angeli JP, Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91-98
202. Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478(3):1338-1343
203. Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther. 2020;5(1):108
204. Wu H, Liu A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res. 2021;49(3):300060521996183
205. Kerins MJ, Ooi A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid Redox Signal. 2018;29(17):1756-1773
206. Zheng ZG, Xu H, Suo SS, Xu XL, Ni MW, Gu LH, Chen W, Wang LY, Zhao Y, Tian B, Hua YJ. The Essential Role of H19 Contributing to Cisplatin Resistance by Regulating Glutathione Metabolism in High-Grade Serous Ovarian Cancer. Sci Rep. 2016;6:26093
207. Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X, Shi L. lncRNA KRAL reverses 5-fluorouracil resistance in hepatocellular carcinoma cells by acting as a ceRNA against miR-141. Cell Commun Signal. 2018;16(1):47
208. Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio MEG, Romeo E, Raimondi L, Caracciolo D, Zuccala V, Rossi M, Neri A, Munshi NC, Tagliaferri P, Tassone P. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018;32(9):1948-1957
209. Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G. Ferroptosis. past, present and future. Cell Death Dis. 2020;11(2):88
210. Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10(1):9-17
211. Reichert CO, de Freitas FA, Sampaio-Silva J, Rokita-Rosa L, Barros PL, Levy D, Bydlowski SP. Ferroptosis Mechanisms Involved in Neurodegenerative Diseases. Int J Mol Sci. 2020 21(22)
212. Xu Y, Luo X, He W, Chen G, Li Y, Li W, Wang X, Lai Y, Ye Y. Long Non-Coding RNA PVT1/miR-150/ HIG2 Axis Regulates the Proliferation, Invasion and the Balance of Iron Metabolism of Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;49(4):1403-1419
213. Hagist S, Sultmann H, Millonig G, Hebling U, Kieslich D, Kuner R, Balaguer S, Seitz HK, Poustka A, Mueller S. In vitro-targeted gene identification in patients with hepatitis C using a genome-wide microarray technology. Hepatology. 2009;49(2):378-386
214. Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang W, Lv S, Li W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020;11(9):751
215. Lo M, Wang YZ, Gout PW. The x(c)- cystine/glutamate antiporter. a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008;215(3):593-602
216. Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, Shi Y, Shen Y, Liu X, Lai W, Yang R, Xiao D, Cheng Y, Liu S, Zhou H, Cao Y, Yu W, Muegge K, Yu H, Tao Y. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Research. 2018;78(13):3484-3496
217. Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57-62
218. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461
219. Xu J, Su C, Zhao F, Tao J, Hu D, Shi A, Pan J, Zhang Y. Paclitaxel promotes lung cancer cell apoptosis via MEG3-P53 pathway activation. Biochem Biophys Res Commun. 2018;504(1):123-128
220. Wang Z, Chen X, Liu N, Shi Y, Liu Y, Ouyang L, Tam S, Xiao D, Liu S, Wen F, Tao Y. A Nuclear Long Non-Coding RNA LINC00618 Accelerates Ferroptosis in a Manner Dependent upon Apoptosis. Mol Ther. 2021;29(1):263-274
221. Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, Shi Y, Chen L, Xiao D, Yu F, Wang X, Zhou H, Cao Y, Liu S, Yan Q, Tao Y, Zhang B. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26(11):2329-2343
222. Werge MP, McCann A, Galsgaard ED, Holst D, Bugge A, Albrechtsen NJW, Gluud LL. The Role of the Transsulfuration Pathway in Non-Alcoholic Fatty Liver Disease. J Clin Med. 2021 10(5)
223. Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23(2):270-278
224. Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, Xu S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. 2019;9(1):16185
225. Chen L, Yang H, Yi Z, Jiang L, Li Y, Han Q, Yang Y, Zhang Q, Yang Z, Kuang Y, Zhu Y. LncRNA GAS5 regulates redox balance and dysregulates the cell cycle and apoptosis in malignant melanoma cells. J Cancer Res Clin Oncol. 2019;145(3):637-652
226. Spencer NY, Yan Z, Boudreau RL, Zhang Y, Luo M, Li Q, Tian X, Shah AM, Davisson RL, Davidson B, Banfi B, Engelhardt JF. Control of hepatic nuclear superoxide production by glucose 6-phosphate dehydrogenase and NADPH oxidase-4. J Biol Chem. 2011;286(11):8977-8987
Author contact
![]() Corresponding authors: Li Cong (E-mail: congliedu.cn); Yiqun Jiang (E-mail: jiangyiqunedu.cn).
Corresponding authors: Li Cong (E-mail: congliedu.cn); Yiqun Jiang (E-mail: jiangyiqunedu.cn).

 Global reach, higher impact
Global reach, higher impact