10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(15):5624-5640. doi:10.7150/ijbs.75994 This issue Cite
Research Paper
Regulation of Podocyte Injury by CircHIPK3/FUS Complex in Diabetic Kidney Disease
1. Department of Nephrology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
2. 5th Medical Department, Medical Faculty Mannheim, University of Heidelberg, D-68167 Mannheim, Germany.
3. Tongji School of Pharmacy, Tongji Medical College, Huazhong University of science and Technology, Wuhan 430030, China.
4. Department of Pharmacology, School of Basic Medical Sciences, Shandong University, Jinan 250012, China.
Received 2022-6-10; Accepted 2022-8-16; Published 2022-9-1
Abstract
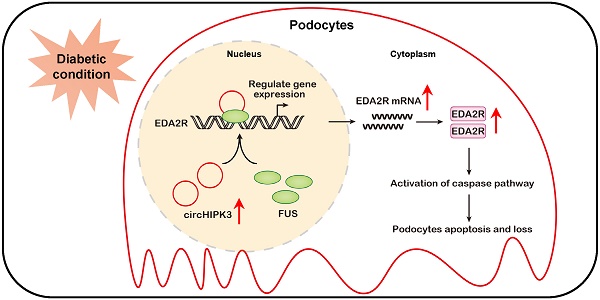
Diabetic kidney disease (DKD) is a major microvascular complication of diabetes mellitus and is one of the leading causes of end-stage kidney disease. Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs that play important roles in various diseases, yet their roles in DKD are poorly understood. CircRNA HIPK3 (circHIPK3), a highly conserved circRNA, is closely related to various cellular functions, including cell proliferation and apoptosis. The association between circHIPK3 and diabetic complications has been well demonstrated in multiple previous studies. However, the role of circHIPK3 in podocyte injury in DKD remains unclear. Herein, we discovered that circHIPK3 expression is markedly elevated in cultured podocytes under high-glucose (HG) conditions and glomeruli of diabetic mice, which is closely associated with podocyte injury in DKD. Functionally, lentivirus-mediated knockdown of circHIPK3 dramatically suppresses HG-induced podocyte apoptosis in vitro. Therapeutically, silencing circHIPK3 by adeno-associated virus-mediated RNA interference ameliorates podocyte injury and albuminuria in STZ-induced diabetic mice. Mechanistically, circHIPK3 facilitates the enrichment of fused in sarcoma (FUS) on the ectodysplasin A2 receptor (EDA2R) promoter, resulting in the upregulation of EDA2R expression and activation of apoptotic signaling. Taken together, these results indicate circHIPK3/FUS/EDA2R axis as a therapeutic target for podocyte injury and DKD progression.
Keywords: podocyte injury, diabetic kidney diseases, circHIPK3, FUS, EDA2R
Introduction
Diabetic kidney disease (DKD), occurring in ~40% of patients with diabetes, is one of the most common and devastating diabetic microvascular complications, which leads to impaired renal function [1, 2]. Although current treatment strategies (intensive glucose control and anti-hypertensive therapies) have meaningfully improved outcomes for diabetes complications, DKD still poses a major risk factor for the progression towards end-stage kidney disease [1]. Abundant evidence indicates that podocyte injury is a critical risk factor for progression of DKD, leading to proteinuria and further kidney damage [3, 4]. Thus, identifying key molecules involved in podocyte injury is of great importance for the development of novel therapeutic strategies for the treatment of patients with DKD. However, to date, the precise molecular mechanisms contributing to podocyte injury and development of DKD have yet to be elucidated. Therefore, effective therapeutic strategies aimed at attenuating or preventing podocyte injury exert the potential to bring huge clinical benefits and economic profits to global health systems.
Previous studies have demonstrated that non-coding RNAs (ncRNAs), such as Tug1 [5, 6], miR-93 [7, 8], and miR-21 [8, 9], contribute to the development and progression of podocyte injury in DKD. CircRNAs are a novel class of endogenous expressed ncRNAs with covalently closed structure and single-stranded that arise from exons or introns, which can regulate gene expression by acting as miRNA sponges, transcription regulators, or RNA-binding protein partners [10, 11]. Previous studies have demonstrated that dysregulated expression of circRNAs is associated with multiple complicated diseases [10-13], including diabetic microvascular complications. For example, circular RNA HIPK3 (circHIPK3) [14], circular RNA PWWP2A (cPWWP2A) [15], and circular RNA ZNF532 (circRNA-ZNF532) [16] have been discovered to play important roles in the development of diabetes-induced retinal vascular leakage. Notably, it has been shown that circRNA.4614 can be considered as potential therapeutic target for diabetic neuropathic pain [17]. Likewise, other studies have also demonstrated that circRNA_010383 [18], and circ_DLGAP4 [19] play critical roles in the initiation and progression of renal fibrosis in DKD. However, to date, little research has been conducted on the potential implications of circRNAs in diabetic podocyte injury. Therefore, more dysregulated circRNAs need to be identified for exploring molecular mechanisms associated with podocyte injury in DKD progression, which is anticipated to make a potential breakthrough in the clinical application for the treatment of DKD.
CircHIPK3, a highly conserved circRNA across different species, is generated from the circularization of HIPK3 gene exon 2, which is expressed ubiquitously in mammalian tissues. Emerging studies have demonstrated that dysregulation of circHIPK3 is associated (perhaps causally) with numerous disease states [20], including multiple diabetes-related complications. However, so far, rare reports have addressed the role of circHIPK3 in the pathogenesis of podocyte injury under diabetic conditions along with its implication on DKD progression. In this study, we identify circHIPK3 upregulation as a key driver of podocyte injury under diabetic conditions, which can promote podocyte loss and DKD progression. Mechanistically, circHIPK3 facilitates the enrichment of fused in sarcoma (FUS) on the ectodysplasin A2 receptor (EDA2R) promoter, resulting in the upregulation of EDA2R expression and activation of apoptotic signaling. In vivo administration of adeno-associated virus-mediated short hairpin RNA (AAV-shRNA) targeting circHIPK3 significantly ameliorates podocyte injury and decreases proteinuria in diabetic mice, indicating the pivotal roles of circHIPK3/FUS/EDA2R axis in DKD progression.
Methods
Cell culture
The conditionally immortalized human podocyte cell line (HPC) was kindly provided by Dr. Heping Ma (Emory University) and the conditionally immortalized mouse podocyte cell line (MPC-5) was obtained from American Type Culture Collection. HPC cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; 10099-141, Gibco, Australia) and MPC-5 cells were propagated in DMEM medium supplemented with 10% FBS as previously described [21, 22]. All cell lines have been authenticated by STR profiling and tested negative for mycoplasma contamination during the period of this study. When the podocytes reached approximately 70%-80% confluence, the cells were maintained at 37 °C for 10-14 days to induce differentiation and then were used for subsequent experiments. Hsa_circ_0000284 overexpression lentivirus and an empty lentivirus, as well as hsa_circ_0000284/mmu_circ_0001052 shRNA lentiviral particle and the indicated scramble shRNA lentiviral were obtained from GeneChem (Shanghai, China). Podocytes were transfected with lentivirus to overexpress or interfere circHIPK3 expression according to the manufacturer's instructions.
RNA isolation and quantitative Real Time PCR (qRT-PCR)
Total RNAs were isolated from cultured cells and mouse glomeruli with FastPure Cell/Tissue Total RNA Isolation Kit (RC112-01, Vazyme, Nanjing) according to directions of the manufacturer. qRT-PCR (Q311-02, Vazyme, Nanjing) was conducted with the StepOne Plus qRT-PCR System (Thermo Fischer Scientific). The specific primers sequences used in this study are listed in Supplementary Table 1.
RNase R and actinomycin D treatment
RNase R (R7092M, Beyotime, China) and actinomycin D (HY-17559, MCE, China) treatment assays were conducted as previously illustrated [14, 18].
RNA fluorescence in situ hybridization (RNA-FISH)
Cy3-labeled circHIPK3, 18s, and U6 probes were acquired from RiboBio (Guangzhou, China). The signal intensity of cy3-labeled probes was detected by the Fluorescent in situ Hybridization Kit (C10910, RiboBio, China) according to directions of the manufacturer. All fluorescence images were captured using Nikon-A1Si Laser Scanning Confocal Microscope (Nikon, Japan).
Nuclear and cytoplasmic extraction
Subcellular fractionation of HPC cells was performed using Cytoplasmic & Nuclear RNA Purification Kit (21000, Norgen Biotek, Canada). Total RNAs were subsequently extracted from both nuclear and cytoplasmic fractions. Then equal volumes of RNA from both fractions were subjected to qRT-PCR (Q311-02, Vazyme, Nanjing) analysis.
RNA pull-down
The biotinylated DNA probes-binding complex was precipitated by incubating streptavidin-coated magnetic beads (M-280, Invitrogen) with cell lysates according to the manufacturer's operating instructions. The relative circHIPK3 enrichment in the sediment was detected by qRT-PCR assay. The binding proteins were subsequently eluted and separated by SDS-PAGE gel (SW109-01, Sevenbio, Beijing, China) electrophoresis, and followed by Coomassie blue staining (P1305, Solarbio, Beijing) or Western blot.
Immunoblot analysis
Total protein extracted from cultured cells or mouse glomeruli was subjected to electrophoresis on SDS-PAGE gels (Sevenbio, Beijing, China) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Missouri, USA). After blocking, the membranes were probed with primary and the corresponding secondary antibodies. Then, immunoreactive bands were visualized by Omni-ECL Enhanced Pico Light Chemiluminescence kit (SQ101, Epizyme Biotech, China). Primary antibodies included: Desmin (ab32362, abcam), IGF2BP1 (ab184305, abcam), IGF2BP3 (ab225697, abcam), CORO1A (ab203698, abcam), BAX (50599-2-Ig, Proteintech), DYKDDDDK Tag (20543-1-AP, Proteintech), IGF2BP2 (11601-1-AP, Proteintech), FUS (11570-1-AP, Proteintech), Cleaved Caspase-3 (Asp175, R&D), Polyclonal anti-podocin (p0372, Sigma-Aldrich), and EDA2R (sc-377423, Santa).
Cross-linking RNA immunoprecipitation (RIP)
Cultured cells were cross-linked at 200 mJ/cm2 using 254 nm UV light with an Ultraviolet Crosslinker (UVP) before being lysed as previously described [23]. RIP assays were conducted using the Magna RIP Kit (17-700, Millipore, MA), with antibodies specific for FUS (11570-1-AP, Proteintech), IGF2BP1 (ab184305, abcam), IGF2BP2 (11601-1-AP, Proteintech), IGF2BP3 (ab225697, abcam), or DYKDDDDK Tag (20543-1-AP, Proteintech). The precipitated RNAs were then analyzed by qRT-PCR. The relative amount of immunoprecipitated RNA in each sample was represented as that of negative (IgG) sample.
In vitro binding assay
A series of FUS truncates were amplified with primer sets (Supplementary Table 1) and subcloned into pCMV 3xFLAG plasmid (Addgene). The FUS-circRNA complexes were pulled down using Anti-Flag magnetic beads (HY-K0207, MCE, China). Protein was measured by Western blot and circRNA expression was detected by qRT-PCR with specific divergent primers (Supplementary Table 1).
Chromatin immunoprecipitation (ChIP)
ChIP assay was conducted using the ChIP Assay kit (P2078, Beyotime, China) according to the manual instruction. Chromatin samples were immunoprecipitated with antibodies against FUS (11570-1-AP, Proteintech) or a negative control Rabbit IgG antibody (31235, Thermo Fisher Scientific). After purification of DNA, gene promoter specific sequences were amplified by qRT-PCR with specific primers. Data were calculated based on fold enrichment of FUS pull-down versus IgG pull-down.
RNA sequencing (RNA-Seq)
RNA sequencing (RNA-seq) experiments were conducted by Seqhealth Technology Co. (Wuhan, China). Briefly, total RNAs were extracted from duplicate of HPC cells using the Total RNA Isolation Kit (Vazyme). Differentially expressed mRNAs between groups were identified with an adjust P-value cutoff of 0.05 and an inter-group absolute fold-change cutoff of 1.5. Differential gene expression information is listed in Supplementary Table 3. Raw data were deposited in the Gene Expression Omnibus database (GSE194107).
Flow cytometric analysis
Podocyte apoptosis rates were examined using a commercial Annexin V-PE/7-AAD Apoptosis Detection Kit (A213-02, Vazyme, Nanjing) according to the manufacturer's instruction and analyzed by flow cytometer (BD Bioscience).
Dual-luciferase reporter assay
The wild-type and the FUS binding site (-569/-562) mutation of human EDA2R promoter (-2,000/-1) were synthesized by AuGCT DNA-SYN Biotechnology Co. Ltd. (Beijing, China) and inserted into the upstream of the firefly luciferase gene in pGL3-Basic luciferase reporter vector (Promega, USA). Luciferase activities were detected by using the Dual-Luciferase® Reporter Assay System kit (E1910, Promega, USA) according to the manufacturer's protocol.
Human renal biopsy samples
Kidney biopsy samples from patients with biopsy-proven DKD were collected at Union Hospital of Tongji Medical College of Huazhong University of Science and Technology (HUST). DKD diagnosis was verified by kidney histological and transmission electron microscopy analysis. Renal samples from patients who received radical nephrectomy for renal carcinoma and without diabetes or kidney disease were collected as control. All specimens were obtained with appropriate informed consent from the patients and approved by the Institutional Review Board of Tongji Medical College of HUST.
Animal studies
All animals were kept in the Specific Pathogen Free (SPF) Laboratory Animal Center of Huazhong University of Science and Technology according to NIH Guidelines for the Care and Use of Laboratory Animals (Approval number: [2021] 2611). Eight-week-old C57BL/6J male mice (GemPharmatech Co.) were treated with an intraperitoneal injection of streptozotocin (150 mg/kg) [24, 25]. One week after streptozotocin injection, mice with blood glucose >16.7 mmol/L were included in the experiments. The recombinant AAV carrying a shRNA against mmu_circ_0001052 (AAV-shRNA) or a negative control shRNA (AAV-scram) were constructed by OBiO Technology (Shanghai, China). After two months of establishment of the DKD model, AAV (2^1011 pfu/per mouse) were delivered by tail vein injection [26, 27]. All mice were killed at 12 weeks after the first injection of AAV. Blood, urine, and kidney tissue specimens were collected for histological and biochemical analyses.
Glomeruli isolation
Mouse glomeruli were isolated according to our previously described [25, 28]. Briefly, mice were anesthetized and the distal abdominal aorta and inferior vena cava, superior mesenteric artery and celiac artery were surgically ligated. After kidneys were perfused with 4^107 inactivated Dynabeads (M-450, Invitrogen) diluted in 10 mL Hank's Balanced Salt Solution (preheated at 37 °C), kidneys were removed immediately and cut into small pieces on ice. Then the minced kidney tissue was digested at 37 °C for 15 mins in a corresponding digestion buffer (consist of 50 U/ml DNase I, 300 U/ml collagenase type II, 1 mg/ml proteinase E). The digested tissue was subsequently lightly filtered through a 100 μm cell strainer, followed by centrifugation at 1500 rpm. After strict washing, the isolated glomeruli were used for RNA isolation and subsequent qRT-PCR analysis as well as protein extraction and Western blot analysis.
Transmission electron microscopy analysis
The handling and examination of electron microscopic samples (mouse kidneys) were performed by the Wuhan Institute of Virology (WIV) of the Chinese Academy of Science (CAS) as described in our previous study [29].
Statistics
All the data statistical analyses in this study were implemented by using GraphPad Prism 8.0 software (La Jolla, USA) and data were expressed as mean ± SD from at least three independent experiments. Unpaired Student's t tests were used to analyze the difference between two groups. One-way ANOVA test was used for comparisons among multiple groups. Spearman's correlation analysis was used to analyze the correlation between different two genes. All statistical analyses were two-tailed, and a P value of <0.05 was considered statistically significant.
Results
CircHIPK3 is up-regulated in HG-stimulated podocyte and participates in HG-induced podocyte apoptosis
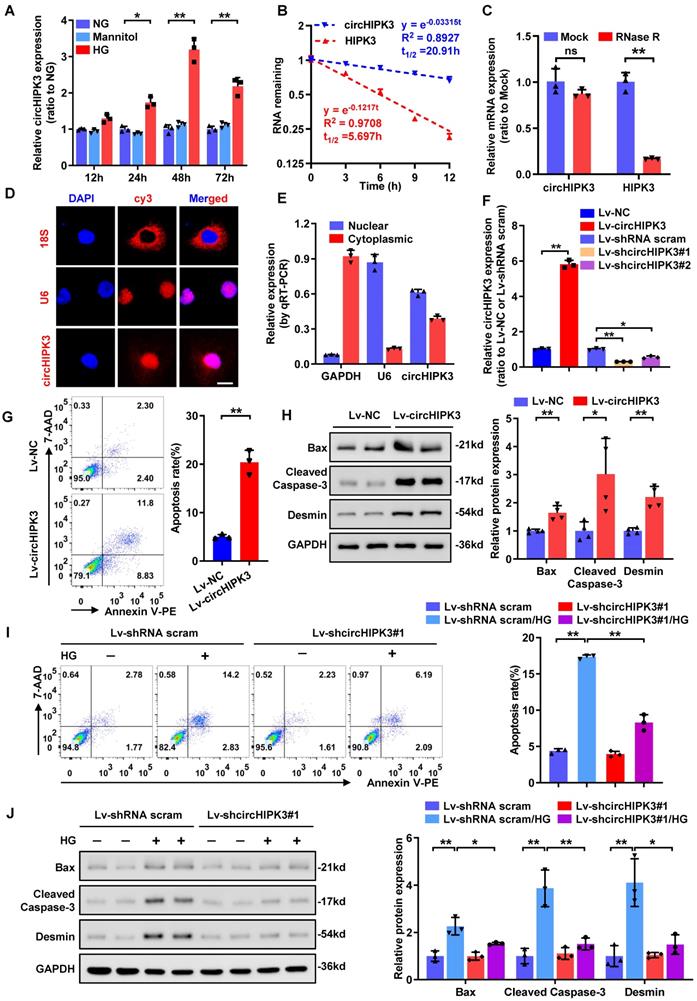
Prior studies have shown that circHIPK3 expression is significantly up-regulated in high-glucose (HG)-treated human retinal endothelial cells and in diabetic mouse retinas [14], implying a potential role of circHIPK3 in diabetic microvascular complications. However, it still remains to be elusive whether circHIPK3 is implicated in podocyte injury in DKD. To determine whether circHIPK3 is involved in HG-induced podocyte injury, we firstly examined the expression pattern of circHIPK3 in HPC cells after HG treatment. qRT-PCR analysis indicated that HG treatment dramatically increased the expression level of circHIPK3 in HPC (Fig. 1A), indicating that circHIPK3 may play an essential role in HG‐induced podocyte injury. Resistant to actinomycin D treatment and RNase R exonuclease digestion confirmed that circHIPK3 exhibited high stability in HPC (Fig. 1B-C). In addition, RNA-FISH detection (Fig. 1D) and qRT-PCR analysis of nuclear and cytoplasmic RNA fractions (Fig. 1E) showed that a large part of circHIPK3 was localized in the nucleus of HPC.
To explore the possible biological function of circHIPK3 in podocytes, lentivirus-mediated circHIPK3 overexpression and knockdown in HPC cells were employed (Fig. 1F). Functional studies revealed that ectopic expression of circHIPK3 resulted in increased podocyte apoptosis (Fig. 1G) and up-regulated expression of pro-apoptosis proteins (Fig. 1H), mimicking the effect of HG stimulation. In contrast, silencing of circHIPK3 decreased cell apoptosis in HG-treated podocyte (Fig. 1I). However, circHIPK3 knockdown did not have a significant effect under normal-glucose conditions (Fig. 1I). These results were further confirmed at protein levels by Western blotting assay (Fig. 1J). Collectively, these findings indicated that circHIPK3 was highly expressed in HG-stimulated podocytes and promoted podocyte injury under diabetic conditions.
CircHIPK3 is up-regulated in HG-stimulated podocyte and participates in HG-induced podocyte injury. A Summarized data of qRT-PCR showing the relative levels of circHIPK3 in HPC treated with 5.5 mmol/L glucose (NG), 40 mmol/L glucose (HG), or mannitol (34.5 mmol/L mannitol plus 5.5 mmol/L glucose) conditions for indicated time. B qRT-PCR assays were conducted to detect the amount of circHIPK3 and HIPK3 mRNA in podocytes after actinomycin D treatment. C Total RNAs were digested with RNase R followed by qRT-PCR detection of circHIPK3 and HIPK3 mRNA expression levels. D Identification of circHIPK3 cytoplasmic and nuclear distribution by RNA FISH in HPC using a cy3-labled junction specific antisense probe (red), with the nuclei staining with DAPI (blue). The 18S and U6 were applied as positive controls. Scale bar, 10 μm. E Identification of circHIPK3 cytoplasmic and nuclear distribution by qRT-PCR analysis in HPC. GAPDH and U6 were used as cytoplasm and nuclear control, respectively. F Validation of lentivirus-mediated circHIPK3 overexpression and knockdown efficiencies by qRT-PCR analysis in HPC cells. G HPC cells with different treatments were stained with 7-AAD and Annexin V-PE, and analyzed by flow cytometry to evaluate the apoptosis rate. Quantification of the apoptotic cells was showed at right panel (n=3). H Representative Western blot gel images (left panel) and summarized data (right panel) showing the relative protein levels of Cleaved Caspase-3, Bax, as well as Desmin in podocytes with different treatments. GAPDH served as loading control (n=4). I The effect of circHIPK3 knockdown on HG-induced HPC apoptosis and the quantification data (n=3). J Representative Western blot gel images (left panel) and summarized data showing the relative protein levels of the indicated proteins in HPC with different treatments. GAPDH served as loading control (n=3). Data are expressed as mean ± SD of three or four independent experiments. One-way ANOVA was used for comparison among multiple groups. Student's t-test was employed for comparisons between two groups. *P < 0.05; **P < 0.01; ns means no significant.
CircHIPK3 interacts with FUS protein in human podocyte
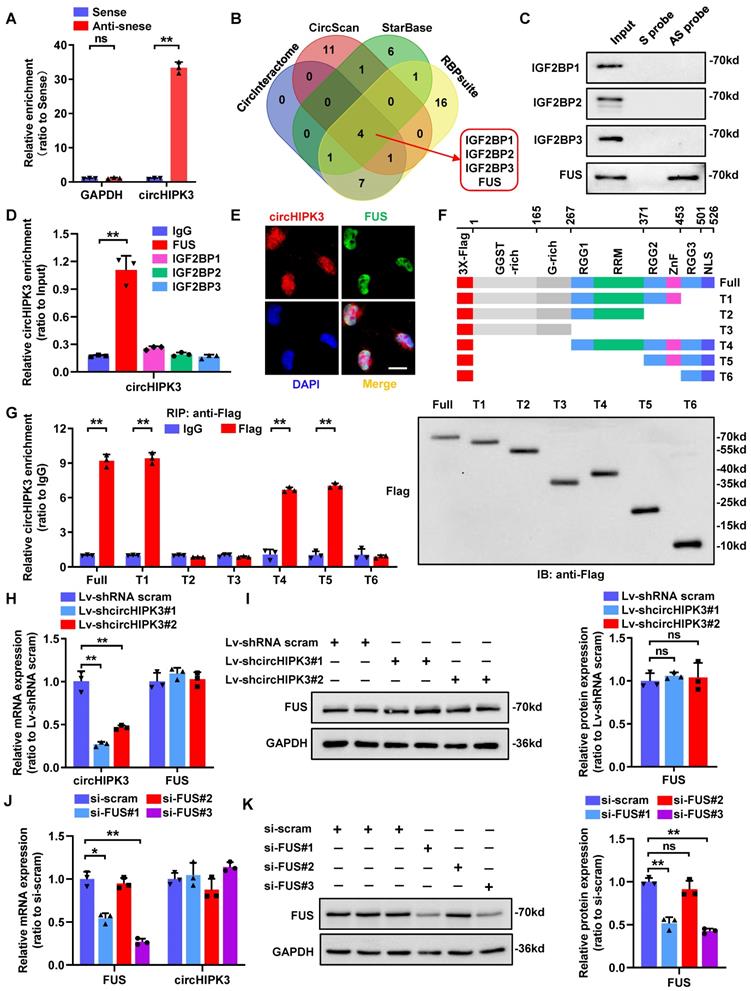
Besides their function as miRNA sponges, circRNAs can exert their biological functions by interacting with RNA-binding proteins [30]. Given that large part of circHIPK3 locates in podocyte nucleus, we next conducted RNA pull-down assay to investigate its protein binding role (Fig. 2A). Then the precipitates were separated by SDS-PAGE followed by Coomassie bright blue staining, and a specific band of ~70 kD was detected in anti-sense probe extracts (Supplementary Fig. 1A). To further explore the potential binding proteins, we performed an in-silico analysis using four bioinformatic website tools (CircInteractome, CircScan, RBPsuite, and StarBase) to predict proteins that may bind to circHIPK3. Among the candidate proteins, four proteins (IGF2BP1/2/3, and FUS) were predicted by all four databases (Fig. 2B; Supplementary Table 2), which was further identified to be FUS by RNA pull-down assay followed by Western blotting (Fig. 2C) and RNA immunoprecipitation (RIP) assay (Supplementary Fig. 1B; Fig. 2D) verification. We further confirmed the co-localization of endogenously expressed circHIPK3 and FUS in the nucleus of podocyte by performing RNA-FISH combined with immunofluorescence (Fig. 2E).
Studies have shown that the C-terminal region of FUS protein, consisting of an RNA recognition motif (RRM) and a zinc finger (ZnF) domain, is an RNA binding motif [31]. To explore whether the two globular domains of FUS are essential for interactions between FUS and circHIPK3, we constructed a series of FUS truncations (Fig. 2F). In vitro binding assays showed that deletion of ZnF (372-453 amino acids), but not the RRM (268-371 amino acids), significantly impaired the interaction between FUS and circHIPK3 (Fig. 2G), suggesting that the ZnF domain was the key region involved in the circHIPK3-FUS interaction. In addition, knockdown of circHIPK3 did not affect FUS expression (Fig. 2H-I), nor did silencing FUS alter the expression level of circHIPK3 (Fig. 2J-K), indicating that their expression levels were independently regulated in HPC. Taken together, our results proposed that circHIPK3/FUS formed an RNA-protein complex through the ZnF domain of FUS in HPC.
CircHIPK3 promotes podocyte damage by upregulating EDA2R
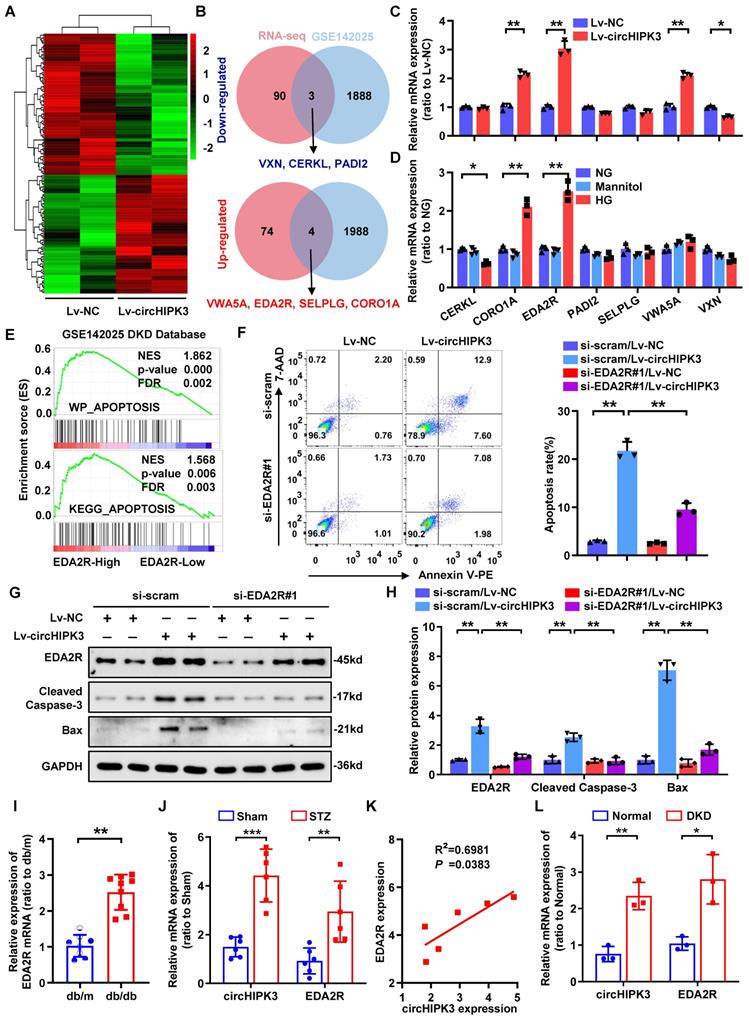
Next, we analyzed potential downstream targets of the increased expression of circHIPK3 accounting for HG-induced podocyte injury. RNA-seq assay revealed 78 up-regulated and 93 down-regulated genes (Fold change > 1.5 or < -1.5, and adjust P-value < 0.05) in HPC upon circHIPK3 overexpression (Fig. 3A; Supplementary Table 3). To identify essential regulators in podocyte injury associated with DKD, we performed a comprehensive analysis of the public DKD datasets (GSE142025) and identified 1992 up-regulated and 1891 down-regulated genes in DKD specimens. Combined analysis of our RNA-seq data and GSE142025, we finally identified 7 differentially expressed genes that are likely to be associated with podocyte injury in DKD (Fig. 3B), but only 4 genes were significantly different in subsequent qRT-PCR verification (Fig. 3C). We speculate that this inconsistency is caused by the low expression abundance of the gene itself, or it may be caused by the interference of homologous gene expression [32, 33]. Interestingly, the expression of ectodysplasin A2 receptor (EDA2R) and coronin 1A (CORO1A) were consistently upregulated upon circHIPK3 overexpression or HG stimulation (Fig. 3C-D; Supplementary Fig. 2A-B). Of note, gene set enrichment analysis (GSEA) indicated that EDA2R (Fig. 3E), but not CORO1A (Supplementary Fig. 2C), was highly correlated with apoptosis-associated pathways based on the data from GSE142025. To further confirm whether circHIPK3-induced podocyte injury via EDA2R or CORO1A, we conducted a series of rescue experiments. The results showed that circHIPK3 induced increased podocytes apoptosis were significantly diminished only in EDA2R-deficient podocyte (Supplementary Fig. 2D-F; Fig. 3F), suggesting that EDA2R mediated circHIPK3-induced podocyte injury. Moreover, EDA2R silencing reversed Caspase-3 activation and EDA2R upregulation in circHIPK3-overexpressed podocytes (Fig. 3G-H). Hence, EDA2R is considered as an important candidate target of circHIPK3.
CircHIPK3 could interact with FUS in podocyte nucleus. A HPC lysates were subjected to RNA pull-down assay. The expression of circHIPK3 was tested by qRT-PCR. B Bioinformatics analysis indicates that IGF2BP1, IGF2BP2, IGF2BP3 and FUS are likely to interact with circHIPK3. C Biotin-labeled RNA pull-down and Western blot assays showing protein pulled down by circHIPK3 from HPC lysates. D RIP assays using anti-IGF2BP1, IGF2BP2, IGF2BP3, FUS or IgG control antibodies and qRT-PCR assays showing the interaction between circHIPK3 and above proteins in HPC cells. E Dual RNA-FISH and immunofluorescence staining assay indicating the colocalization of circHIPK3 (red) and FUS (green) in HPC cells. F Schematic diagram revealing the domains of FUS truncations. G In vitro binding assay depicting the recovered circHIPK3 levels detected by qRT-PCR (left panel) after incubation with full-length or truncation forms of Flag-tagged recombinant FUS protein validated by Western blot (right panel). H, I Expression of FUS and circHIPK3 was confirmed by qRT-PCR (H) or Western blot (I) in HPC transfected with Lv-circHIPK3 shRNAs (n=3). J, K Expression of circHIPK3 and FUS were verified by qRT-PCR (J) or Western blot (K) in HPC transfected with FUS siRNAs (n=3). Data are shown as mean ± SD of three independent experiments. Student's t-test was employed for comparisons between two groups. *P < 0.05, **P < 0.01; ns represents no significant.
In addition, EDA2R was significantly increased in micro-dissected glomeruli from subjects with db/db mice based on a publicly available data set from Nephroseq (https://www.nephroseq.org/) (Fig. 3I). From the same database, higher expression levels of EDA2R were positively correlated with increased levels of urinary albumin/creatinine ratio (UACR) in db/db mice (Supplementary Fig. 2G). Consistently, our animal studies indicated that EDA2R expression was significant up-regulated in glomeruli from STZ-induced diabetic mice as compared with non-diabetic mice (Fig. 3J; Supplementary Fig. 2H-I). Meanwhile, analysis of the correlation between EDA2R and circHIPK3 transcriptional levels showed that EDA2R was positively correlated with circHIPK3 expression in glomeruli micro-dissected from diabetic kidneys (Fig. 3K). These data were confirmed in renal biopsy samples from patients with biopsy-proven DKD (Fig. 3L). From these findings, we concluded that circHIPK3-induced podocyte injury probably mainly through upregulating EDA2R expression.
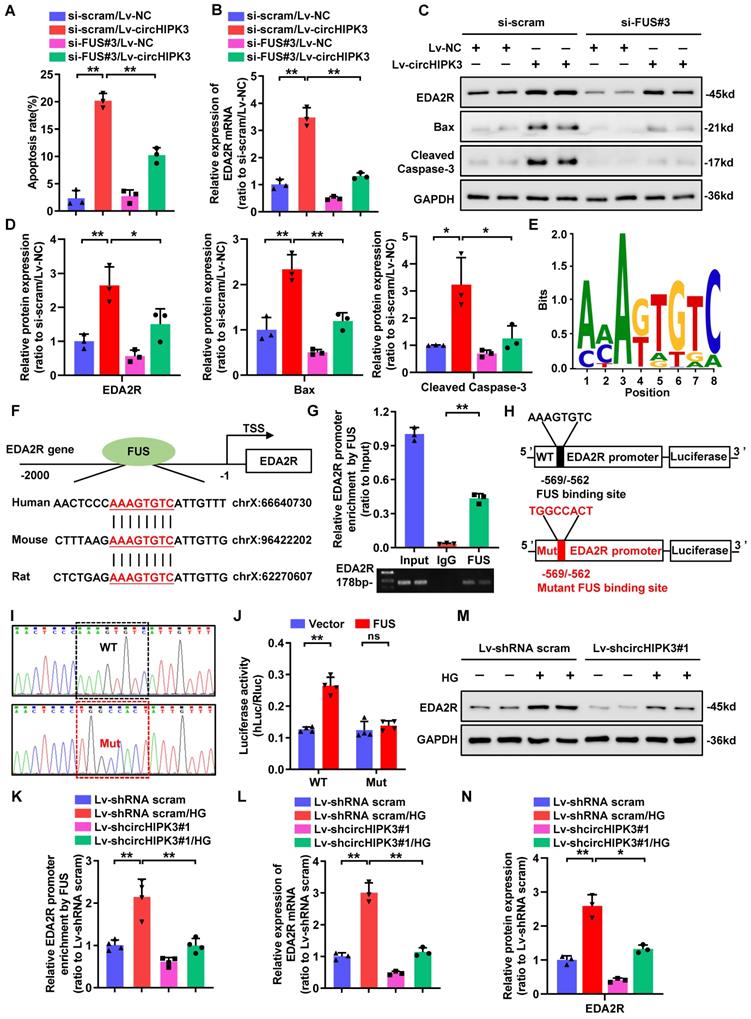
CircHIPK3 facilitates the expression of EDA2R through FUS
We then returned to consider how circHIPK3 regulated the expression of EDA2R. Previous studies have confirmed that circRNAs located in the nucleus are mainly involved in transcriptional regulation through interacting with DNA-binding proteins or recruiting transcription cofactors [11-13]. To investigate the role of circHIPK3/FUS complex in regulating EDA2R expression, we first performed FUS knockdown in circHIPK3-overexpressed HPC. Flow cytometric analysis showed that silencing of FUS significantly inhibited circHIPK3-induced HPC apoptosis (Fig. 4A). Furthermore, knockdown of FUS suppressed circHIPK3-induced upregulation of EDA2R at both mRNA and protein levels (Fig. 4B-C), as well as increased Cleaved Caspase-3 and Bax protein expression (Fig. 4C-D), suggesting that circHIPK3-induced podocytes apoptosis and upregulation of EDA2R were largely dependent on FUS. Considering that FUS could regulate transcription in the nucleus [34, 35], we speculated that circHIPK3 may regulate EDA2R expression through interacting with FUS at transcription level. To verify this hypothesis, we scanned the DNA sequence of EDA2R promoter for putative FUS binding sites. Interestingly, one highly conserved putative binding site for FUS was identified in EDA2R promoter DNA sequence (Fig. 4E-F). To verify the binding site, a ChIP assay was performed with PCR primers flanking the region of the FUS binding site and found that FUS could specifically bind to EDA2R promoter region (Fig. 4G). To further verify whether EDA2R gene was the direct target of FUS, a dual-luciferase reporter system was used. A firefly luciferase reporter construct containing the wild-type (WT) or mutant (Mut) EDA2R promoter was co-transfected with FUS overexpression plasmid or empty plasmid (Fig. 4H-I). Luciferase reporter assays showed that ectopic expression of FUS significantly enhanced the luciferase activity of the WT EDA2R promoter reporter, whereas mutation of FUS binding site abolished this effect (Fig. 4J).
To further confirm whether circHIPK3 could recruit FUS on EDA2R promoter, we performed ChIP assay in HPC with or without circHIPK3 knockdown. ChIP analysis revealed that binding of FUS to the EDA2R promoter was strikingly enhanced by HG treatment, while this association was disturbed by circHIPK3 silencing (Fig. 4K). Furthermore, circHIPK3 knockdown reduced HG-induced upregulation of EDA2R at both mRNA and protein levels (Fig. 4L-N). These data suggested that circHIPK3 facilitated the expression of EDA2R and subsequent apoptosis by recruiting FUS to the promoter of EDA2R in HPC.
Knockdown of circHIPK3 in mouse podocyte attenuates HG-induced apoptosis through regulating EDA2R expression
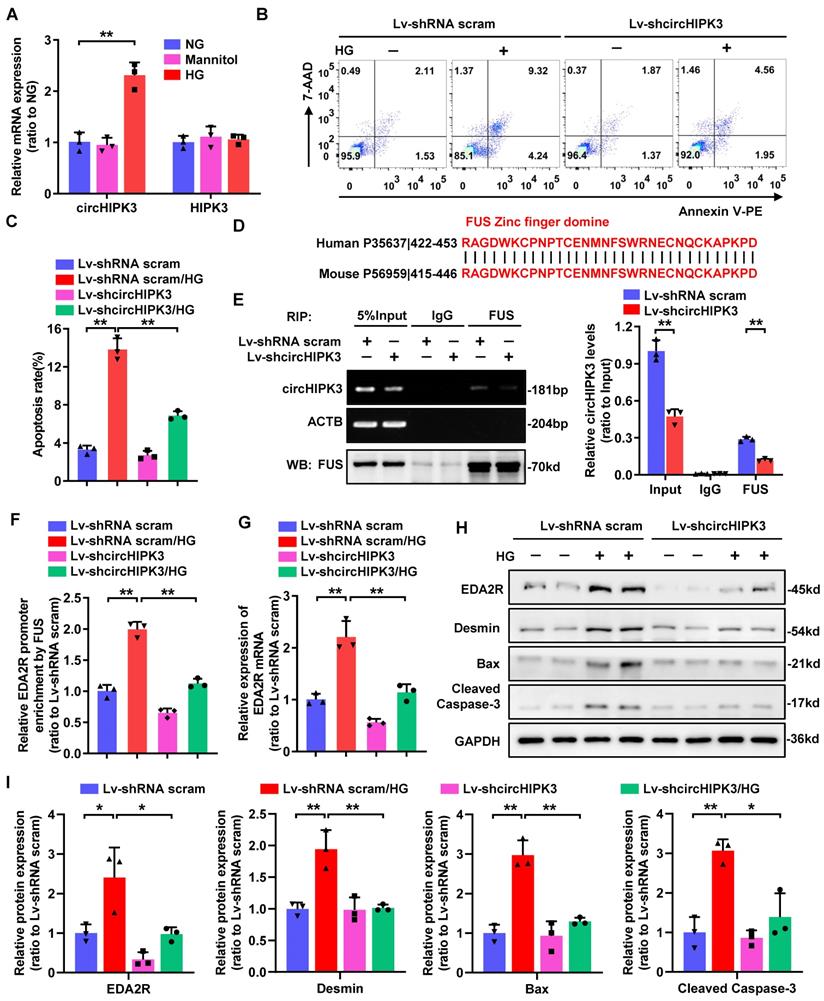
As circHIPK3 is highly conserved between human and mouse, we therefore evaluated whether circHIPK3 is involved in HG-induced mouse podocyte (MPC-5) injury. To delineate the role of circHIPK3 in HG-stimulated MPC-5, we firstly examined its expression pattern. Consistently, circHIPK3 expression was significantly up-regulated in HG-stimulated MPC-5 (Fig. 5A). Further studies revealed that silencing of circHIPK3 attenuated HG-induced MPC-5 apoptosis (Fig. 5B-C), indicating that circHIPK3 is indeed involved in HG-induced mouse podocyte injury. In accordance with the reported that FUS preferential binding to the “UGGU” consensus [31], we identified four conserved potential FUS-binding regions in both human and mouse circHIPK3 sequences (Supplementary Fig. 1C). Moreover, alignment of FUS protein sequence of human and mouse showing the ZnF domain is highly conserved (100% sequence identity) (Fig. 5D), indicating a possible binding between FUS and circHIPK3 in mouse podocyte. A RIP assay confirmed that endogenous FUS could interact with circHIPK3 in MPC-5, which was significantly decreased by silencing of circHIPK3 (Fig. 5E). In addition, circHIPK3 was also confirmed to recruit FUS to EDA2R promoter in MPC-5 with HG treatment (Fig. 5F). Consistently, knockdown of circHIPK3 significantly decreased HG-induced upregulation of EDA2R mRNA and protein levels, as well as increased Cleaved Caspase-3, Bax, and Desmin protein expression (Fig. 5G-I). Altogether, these results demonstrated that knockdown of circHIPK3 alleviated HG-induced MPC-5 injury via decreasing EDA2R expression, which further confirmed that circHIPK3 was a pro-apoptotic factor in podocyte.
CircHIPK3 promotes podocyte damage by upregulating EDA2R. A Clustered heatmap of significant differentially expressed mRNAs in HPC transfected with circHIPK3 overexpression lentivirus versus control lentivirus. Each sample contained a mixture of three repeats. B Schematic flowchart showed the overlapping of circHIPK3-regulated mRNAs identified by RNA-seq in HPC cells and published GSE142025 dataset in human DKD (filtered by fold change > 1.5 or < -1.5 and adjust p-value < 0.05). C Expression levels of indicated mRNAs in HPC cells with circHIPK3 overexpression. D Expression levels of indicated mRNAs in HPC cells under HG (40 mmol/L) stimulation. E Gene set enrichment analysis (GSEA) of GEO datasets (GSE142025) showing that higher EDA2R expression is significantly associated with apoptosis in DKD. F HPC cells with different treatments were stained with Annexin V-PE and 7-AAD, and analyzed by flow cytometry to evaluate cell apoptosis rate. Summarized was showed at right panel (n = 3). G, H Western blot analysis of EDA2R, Bax, and Cleaved Caspase-3 in HPC with different treatments (n=3). I Nephroseq expression data for EDA2R in db/m mice glomeruli (n=8) and db/db mice glomeruli (n=9). J qRT-PCR assays were conducted to detect circHIPK3 and EDA2R expression in STZ-induced diabetic mouse glomeruli (n=6) after five months diabetes induction, in comparison with the control mouse glomeruli (n=6). Expression values were normalized to ACTB mRNA. K Correlation analysis revealed positive correlation between the levels of circHIPK3 and EDA2R mRNA from the diabetic mouse glomeruli. L qRT-PCR analysis of circHIPK3 and EDA2R expression levels in renal biopsy samples from patients with DKD (n = 3 for control, n = 3 for patients with DKD). Data are shown as mean ± SD of three independent experiments. Student's t-test was employed for comparisons between two groups. One-way ANOVA was used for comparison among multiple groups. Correlation between circHIPK3 and EDA2R expression was analyzed by Pearson's correlation. *P < 0.05, **P < 0.01, ***P < 0.001.
CircHIPK3 facilitates the expression of EDA2R through FUS. A Summarized data showing the level of cell apoptosis determined by flow cytometric analysis in podocytes with different treatments (n=3). B qRT-PCR analysis of EDA2R mRNA expression levels in HPC cells transfected with Lv-NC or Lv-circHIPK3 and those co-transfected with si-FUS#3 or si-scram. C, D Western blot analysis of EDA2R, Bax, and Cleaved Caspase-3 in HPC with different treatments (n=3). E The predicted sequence of FUS-binding motif as identified by MatrixREDUCE. F The predicted FUS binding site at the promoter of the evolutionarily conserved region of EDA2R in human, mouse and rat, indicated by red and underlined. G Chromatin immunoprecipitation (ChIP) assays using FUS antibody indicating that FUS could bind to promoter of EDA2R gene in HPC cells. IgG was applied as negative control. H, I Schematic model of mutation (H) and sequencing of mutation (I). J Luciferase activity of the reporter vector containing the wide type (WT) or mutant (Mut) promoter of EDA2R was determined after co-transfection with control or FUS expressing plasmids in HPC cells. K ChIP analysis showed that the interaction between EDA2R gene promoter and FUS protein in HPC cells with different treatments. L qRT-PCR analysis of EDA2R mRNA expression levels in HPC cells with indicated treatments. M, N Representative Western blot gel images (M) and summarized data (N) showing the effect of circHIPK3 knockdown on the protein level of EDA2R in HPC cells treated with HG (40mmol/L) (n=3). Values are expressed as mean ± SD of three or four independent experiments. One-way ANOVA was used for comparison among multiple groups. Student's t-test was employed for comparisons between two groups. *P < 0.05, **P < 0.01; ns represents no significant.
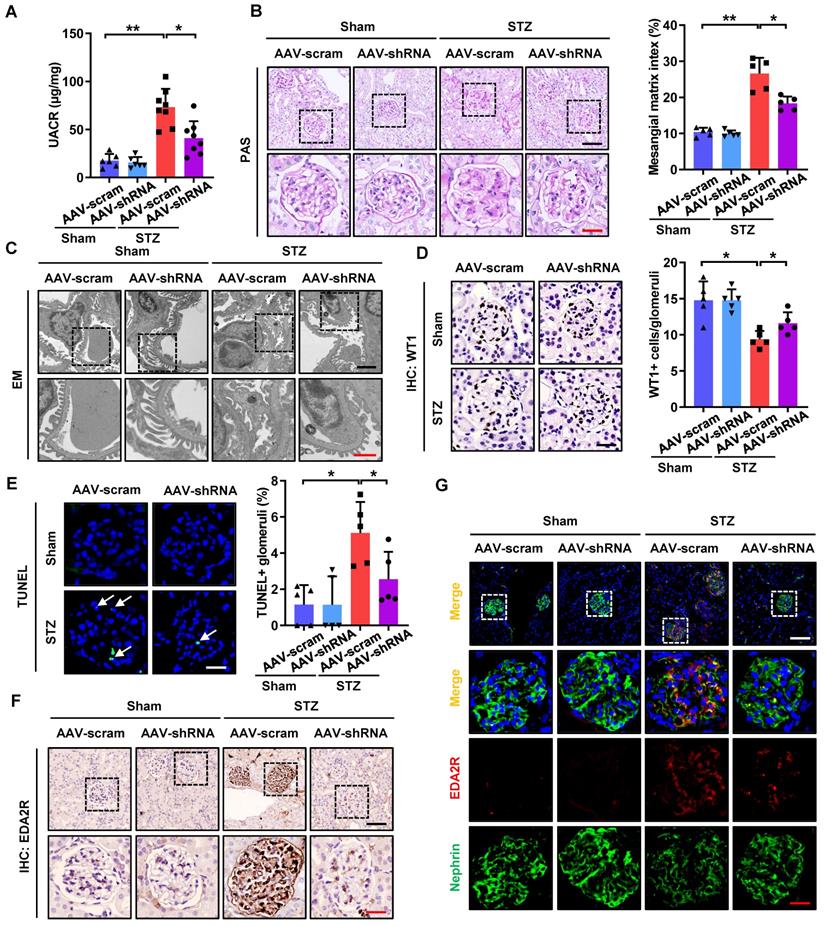
CircHIPK3 knockdown attenuates podocyte injury and DKD in STZ-induced diabetic mice
As silencing of circHIPK3 significantly ameliorated HG-induced podocyte injury in vitro, we next examined the role of circHIPK3 in podocyte injury in vivo. To explore the potential for therapeutic targeting of circHIPK3, we silenced circHIPK3 in STZ-induced diabetic mice with adeno-associated virus (AAV) coated with shRNA-circHIPK3-mCherry (Supplementary Fig. 3A). Our results confirmed the efficiency of circHIPK3 knockdown in vivo, showing that diabetic mice received AAV-shRNA-circHIPK3 (AAV-shRNA) markedly decreased the level of circHIPK3 in isolated glomeruli by qRT-PCR analysis (Supplementary Fig. 3B). Additionally, as shown in Supplementary Fig. 3C, knockdown of circHIPK3 had no significant effects on body weight gain or the changes in blood glucose levels in the diabetic mice, which is consistent with reports from previous studies [14, 36]. To examine the outcome of circHIPK3 depletion in DKD, we further evaluated renal function, urine protein levels, and histological changes in the mice. Our data demonstrated that knockdown of circHIPK3 significantly ameliorated renal injury in STZ-induced diabetic mice as evidenced by reduced albuminuria (Fig. 6A), decreased mesangial expansion (Fig. 6B), as well as attenuated podocyte foot process effacement (Fig. 6C) and podocyte loss (Fig. 6D). To further quantitatively evaluate glomerular cell apoptosis, apoptotic cells accessed by TUNEL staining were evaluated. The results showed that knockdown of circHIPK3 decreased glomerular cell apoptosis in diabetic mice (Fig. 6E). Consistently, depletion of circHIPK3 decreased the protein levels of Cleaved Caspase-3 in glomerular tissues of diabetic mice (Supplementary Fig. 3E-F). Moreover, qRT-PCR analysis (Supplementary Fig. 3D), Western blotting (Supplementary Fig. 3E-F), IHC staining (Fig. 6F), and immunofluorescence (Fig. 6G) results further confirmed that silencing circHIPK3 significantly decreased EDA2R expression and restored the loss of podocyte marker in STZ-induced diabetic mice. Taken together, these data suggested that knockdown of circHIPK3 expression remarkably attenuated podocytes injury and DKD progression in diabetic mice.
Discussion
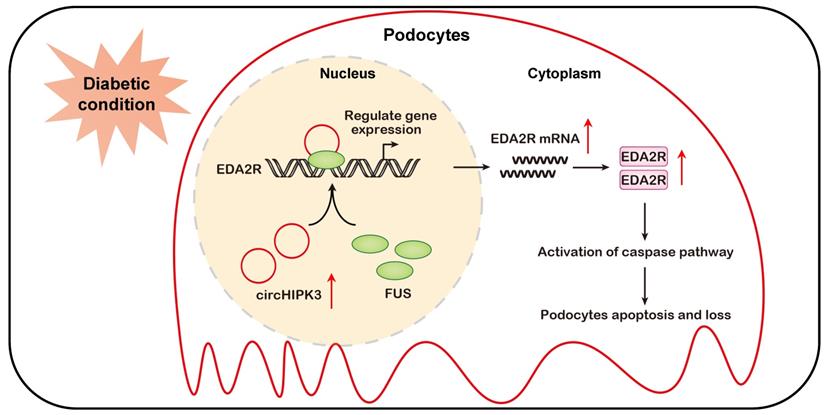
In the present study, we identify circHIPK3, a highly conserved circRNA, as an important signature circRNA in DKD. Our evidence shows that the level of circHIPK3 is increased in podocyte cultured in HG condition and diabetic mice glomerular, and silencing circHIPK3 by adeno-associated virus-mediated RNA interference ameliorates podocyte injury and albuminuria in STZ-induced diabetic mice. Mechanistically, circHIPK3 promotes the enrichment of FUS on EDA2R promoter, resulting in the upregulation of EDA2R expression and activation of apoptotic signaling (Fig. 7). These findings highlight a novel role for circHIPK3 in regulating podocyte injury in DKD, representing a promising therapeutic target for DKD.
CircRNAs function as important regulators in various physiological and pathophysiological process in human health and disease [11]. Emerging studies have demonstrated that circRNAs located in cytoplasmic can function as efficient miRNA sponges to inhibit miRNA-target interactions [10-12]. Alternatively, circRNAs can also facilitate chromatin modifications and promote gene expression in the nucleus. For example, circ-CTNNB1 accelerates tumor progression through DDX3 mediated transactivation of YY1 [23]. In addition, ci-ankrd52 regulates target gene expression by modulating the elongation activity of RNA polymerase II [37]. Previous studies have shown that circHIPK3 mainly function as an endogenous miRNA sponge to regulate the expression of target genes [14, 36, 38, 39]. In our study, we expand the understanding of the mechanisms of circHIPK3 by showing that circHIPK3 can also function via FUS-mediated target gene transcriptional activation.
As a member of the ten-eleven translocation (TET) family, FUS is predominantly expressed in nucleus and involved in numerous cellular processes, including the regulation of transcription and translation [40]. FUS has been identified as a vital modulator of cell apoptosis. In prostate cancer, increased expression of FUS promotes cell death through activation of apoptotic pathways [41]. Meanwhile, ectopic expression of FUS induces neurotoxicity through the mitochondrial apoptotic pathway [42]. While on the contrary, silencing of FUS activates cell apoptosis in gastric cancer [43]. These findings indicate that FUS exerts pro-apoptotic or anti-apoptotic effects in a context-dependent manner. As a transcription factor, FUS has been reported to bind to the promoters of target genes such as COL4A2 [35], CCND1 [41], and MECP2 [44].
Knockdown of circHIPK3 in mouse podocyte attenuates HG-induced apoptosis through regulating EDA2R expression. A Expression levels of circHIPK3 and HIPK3 in MPC-5 cells cultured in media containing 5.5 mmol/L glucose (NG), 40 mmol/L glucose (HG), or mannitol/glucose (34.5 mmol/L mannitol plus 5.5 mmol/L glucose) conditions for 48h. B, C The effects of circHIPK3 knockdown on apoptosis in MPC-5 cells induced by HG treatment, and the quantification data (n=3). D The amino acid sequence of the highly conserved zinc finger (ZnF) domine displaying 100% identity between human and mouse. E RIP (left) and qRT-PCR (right) assays using FUS antibody showing the interaction between circHIPK3 and FUS protein in MPC-5 cells transfected with lentivirus carrying circHIPK3 shRNA or shRNA scram. The IgG-bound RNA was taken as a negative control. F ChIP analysis showing the interaction between EDA2R gene promoter and FUS protein in MPC-5 cells with different treatments. G qRT-PCR showing the expression of EDA2R in MPC-5 cells transfected with Lv-shcircHIPK3 or Lv-shRNA scram under HG (40mmol/L) treatment. Expression values were normalized to ACTB mRNA (n = 3). H, I Representative Western blot gel documents showing the relative protein levels of EDA2R, Desmin, Bax, and Cleaved Caspase-3 in MPC-5 cells with different treatments (n=3). Data are expressed as mean ± SD of three independent experiments. One-way ANOVA was used for comparison among multiple groups. *P < 0.05, **P < 0.01.
In vivo knockdown of circHIPK3 ameliorates renal injury in STZ-induced diabetic mice. A Urinary albumin-to-creatinine ratio (UACR) in different groups of mice was measured. B Representative images of Periodic-acid-Schiff (PAS) staining and mesangial matrix index quantifications (calculated by Image J software) showing typical changes in glomerular structure in different groups of mice. Scale bar: black 50 μm, red 20 μm. C Representative electron microscopy analysis of renal sections in different groups of mice. Scale bar: black 2 μm, red 1 μm. D Representative images of WT1 IHC staining and quantification of WT1-positive glomerular cells showed that knockdown of circHIPK3 restored podocyte loss. Scale bar: 20 μm. E Representative images of TUNEL staining (left panel) and quantification of TUNEL-positive glomeruli (right panel) in renal sections in different groups. Scale bar: 20 μm. White arrow: TUNEL staining of positive cells. F IHC analysis of EDA2R expression in renal tissues in different groups of mice. Scale bar: black 50 μm, red 20 μm. G Representative confocal microscopic images showing the expressions of Nephrin and EDA2R in podocytes of the kidney from different groups of mice. Scale bar: white 50 μm, red 20 μm. Data are expressed as means ± SD. One-way ANOVA was used for comparison among multiple groups (n = 5-8 for each group). *P < 0.05, **P < 0.01.
Schematic depicting circHIPK3 upregulation exacerbates podocyte injuries and proteinuria in DKD through facilitating transcriptional activation of EDA2R in a FUS dependent manner. Under diabetic conditions, upregulated circHIPK3 facilitates the enrichment of FUS on EDA2R promoter, resulting in activation of EDA2R-mediated apoptosis signaling pathway in podocytes.
Recent studies also reveal that circFndc3b plays a protective role in myocardial infarction via interacting with FUS to regulate VEGF expression [45]. However, the role of circRNA in regulating FUS activity in DKD progression remains to be determined. In the current study, our results indicated that circHIPK3 could directly bind to the ZnF domain of FUS protein. We speculate that the binding of circHIPK3 may result in altered FUS protein structure that facilitates its binding to the target gene promoter, with the underlying mechanisms warranting further investigation. Because inhibition of FUS expression abolishes circHIPK3-induced podocyte apoptosis, it is suggested that the pro-apoptotic role of circHIPK3 in podocyte injury under diabetic condition is mediated, at least in part, through interacting with FUS.
RNA-seq and bioinformatics analysis were conducted to screen the downstream target genes of circHIPK3/FUS complex and further indicated that EDA2R expression was regulated by circHIPK3/FUS and mediated circHIPK3-induced podocyte injury in DKD. EDA2R, also known as TNF-RSF27, belongs the tumor necrosis factor receptor superfamily and participates in a variety of signaling pathways including cell apoptosis, immune response, inflammation, and so on [46, 47]. Although EDA2R lacks a discernible death domain, it can induce apoptotic signaling through the activation of the caspase cascade and induction of apoptosis [47-49]. The expression of EDA2R has been reported to be significantly up-regulated in diabetic kidneys (in both type 1 and type 2 diabetic mice) [50, 51]. Moreover, another recent study reveals that EDA2R expression is up-regulated in HG-stimulated podocytes, which mediates podocyte injury through ROS generation [52]. However, the specific mechanisms through which transcriptional regulators modulate EDA2R expression remains unclear. In the current study, mining of public DKD datasets reveals that higher expression of EDA2R is highly associated with apoptosis. Functional studies show that knockdown of EDA2R significantly abolishes circHIPK3-induced podocyte apoptosis, suggesting the pro-apoptotic role of EDA2R in podocyte. Additionally, our results indicate that circHIPK3 directly binds the ZnF domain of FUS to increase its binding to EDA2R promoter, resulting in transcription activation of EDA2R, which provides further evidences for the transcriptional regulation of EDA2R. Furthermore, in vivo knockdown of circHIPK3 by tail vein injection of AAV-shRNA significantly attenuates the upregulation of EDA2R expression in the glomeruli induced by diabetic conditions, indicating that this signaling pathway is likely to be glomerular cell-specific. Nevertheless, the underlying mechanisms warrants further investigation. Besides, it's also proved that circHIPK3 can regulate CORO1A expression, however, CORO1A does not play a major role in circHIPK3-mediated podocyte apoptosis in vitro (Supplementary Figure 2D-E). Several previous studies have shown that CORO1A is a member of an evolutionary conserved actin-associated family of proteins, which were initially identified as important regulators of actin cytoskeleton-dependent processes [53-55], including phagocytosis, cell migration, polarization and cytokinesis. Based on the above results and previous studies, we speculate that CORO1A may be involved in regulating circHIPK3-induced podocyte injury through other ways in DKD, whereas the precise mechanism is still worth further exploring and analyzing.
In summary, to the best of our knowledge, we have demonstrated for the first time, that circHIPK3 is up-regulated in diabetic kidneys and mediates podocyte injury and proteinuria. Mechanistic studies reveal that circHIPK3 cooperates with FUS to promote the podocyte injury and the development of DKD through facilitating transcriptional activation of EDA2R (Fig. 7). Administration of AAV-mediated shRNA targeting circHIPK3 decreases proteinuria, and prevents podocyte damage in DKD. This study extends our knowledge about the transcription regulation of EDA2R signaling by circRNA, suggesting that targeted inhibition of circHIPK3-mediated EDA2R signaling may provide a novel approach for the treatment of DKD podocyte injury.
Abbreviations
DKD: Diabetic kidney disease; circRNA: circular RNA; ncRNA: noncoding RNA; HG: high glucose; FUS: Fused in sarcoma; IGF2BP1/2/3: insulin-like growth factor 2 mRNA-binding protein 1/2/3; EDA2R: ectodysplasin A2 receptor; CORO1A: coronin 1A; HPC: Immortalized human podocyte; MPC: conditionally immortalized mouse podocyte; ChIP: chromatin immunoprecipitation; AAV: Adeno-associated virus.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This work was supported by the National Natural Science Foundation of China grants 81961138007, 81770711, 81974096 (to Pro. C.Z.), 81873602 (to Pro. H.S.), 81700603 (to Dr. H.T.), the Program for HUST Academic Frontier Youth Team (2017QYTD20, to Pro. C.Z.), and the Deutsche Forschungsgemeinschaft (DFG: LI 3384/2-1, to Pro. Hans-Peter Hammes).
Author Contributions
C.Z., H.S. and F.L. designed the study. F.L., J.H., Y.-R.X, Y.-L.C., L.T., and H.T., conducted the experiments and analyzed the data. F.L., and Y.-L.C., developed the methodology (e.g., statistical analysis, biostatistics, computational analysis). C.Z., H.S., F.Y., K.H., H.-P.H., and J.-H.L. interpreted the data and provided conceptual advice. F.L., analyzed and interpreted the data and wrote the paper. C.Z., H.S, F.L., and C.-Y.Z. review, and/or revision of the manuscript. C.Z., H.S., H.T. and H.-P.H. provided research support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Alicic RZ, Rooney MT, Tuttle KR. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clinical journal of the American Society of Nephrology: CJASN. 2017;12:2032-45
2. Alicic RZ, Cox EJ, Neumiller JJ, Tuttle KR. Incretin drugs in diabetic kidney disease: biological mechanisms and clinical evidence. Nat Rev Nephrol. 2021;17:227-44
3. Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124:2333-40
4. Mathieson PW. The podocyte as a target for therapies-new and old. Nat Rev Nephrol. 2011;8:52-6
5. Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL. et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126:4205-18
6. Li L, Long J, Mise K, Galvan DL, Overbeek PA, Tan L. et al. PGC1α is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites. Cell Rep. 2021;36:109510
7. Badal SS, Wang Y, Long J, Corcoran DL, Chang BH, Truong LD. et al. miR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy. Nat Commun. 2016;7:12076
8. Liu F, Chen J, Luo C, Meng X. Pathogenic Role of MicroRNA Dysregulation in Podocytopathies. Front Physiol. 2022;13:948094
9. Kölling M, Kaucsar T, Schauerte C, Hübner A, Dettling A, Park JK. et al. Therapeutic miR-21 Silencing Ameliorates Diabetic Kidney Disease in Mice. Mol Ther. 2017;25:165-80
10. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
11. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475-90
12. Brandenburger T, Salgado Somoza A, Devaux Y, Lorenzen JM. Noncoding RNAs in acute kidney injury. Kidney Int. 2018;94:870-81
13. van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17:814-26
14. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X. et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629-42
15. Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen X. et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc Natl Acad Sci U S A. 2019;116:7455-64
16. Jiang Q, Liu C, Li CP, Xu SS, Yao MD, Ge HM. et al. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J Clin Invest. 2020;130:3833-47
17. Zhang HH, Zhang Y, Wang X, Yang P, Zhang BY, Hu S. et al. Circular RNA profile in diabetic peripheral neuropathy: analysis of coexpression networks of circular RNAs and mRNAs. Epigenomics. 2020;12:843-57
18. Peng F, Gong W, Li S, Yin B, Zhao C, Liu W. et al. circRNA_010383 Acts as a Sponge for miR-135a, and Its Downregulated Expression Contributes to Renal Fibrosis in Diabetic Nephropathy. Diabetes. 2021;70:603-15
19. Bai S, Xiong X, Tang B, Ji T, Li X, Qu X. et al. Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-κB/MMP-2 axis. Cell Death Dis. 2020;11:1008
20. Fu Y, Sun H. Biogenesis, cellular effects, and biomarker value of circHIPK3. Cancer Cell Int. 2021;21:256
21. Su H, Wan Q, Tian XJ, He FF, Gao P, Tang H. et al. MAD2B contributes to podocyte injury of diabetic nephropathy via inducing cyclin B1 and Skp2 accumulation. Am J Physiol Renal Physiol. 2015;308:F728-36
22. Lei CT, Su H, Ye C, Tang H, Gao P, Wan C. et al. The classic signalling and trans-signalling of interleukin-6 are both injurious in podocyte under high glucose exposure. J Cell Mol Med. 2018;22:251-60
23. Yang F, Fang E, Mei H, Chen Y, Li H, Li D. et al. Cis-Acting circ-CTNNB1 Promotes β-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019;79:557-71
24. Qiang G, Xue S, Yang JJ, Du G, Pang X, Li X. et al. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes. 2014;63:1394-409
25. Li MR, Lei CT, Tang H, Yin XJ, Hao Z, Qiu Y. et al. MAD2B promotes podocyte injury through regulating Numb-dependent Notch 1 pathway in diabetic nephropathy. Int J Biol Sci. 2022;18:1896-911
26. Østergaard MV, Secher T, Christensen M, Salinas CG, Roostalu U, Skytte JL. et al. Therapeutic effects of lisinopril and empagliflozin in a mouse model of hypertension-accelerated diabetic kidney disease. Am J Physiol Renal Physiol. 2021;321:F149-f61
27. Sembach FE, Ægidius HM, Fink LN, Secher T, Aarup A, Jelsing J. et al. Integrative transcriptomic profiling of a mouse model of hypertension-accelerated diabetic kidney disease. Disease models & mechanisms. 2021 14
28. Fu Y, Sun Y, Wang M, Hou Y, Huang W, Zhou D. et al. Elevation of JAML Promotes Diabetic Kidney Disease by Modulating Podocyte Lipid Metabolism. Cell Metab. 2020;32:1052-62.e8
29. Bao D, Su H, Lei CT, Tang H, Ye C, Xiong W. et al. MAD2B-mediated cell cycle reentry of podocytes is involved in the pathogenesis of FSGS. Int J Biol Sci. 2021;17:4396-408
30. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402-12
31. Loughlin FE, Lukavsky PJ, Kazeeva T, Reber S, Hock EM, Colombo M. et al. The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity. Mol Cell. 2019;73:490-504.e6
32. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621-8
33. Kang BH, Jensen KJ, Hatch JA, Janes KA. Simultaneous profiling of 194 distinct receptor transcripts in human cells. Science signaling. 2013;6:rs13
34. Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A. et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529
35. Chiusa M, Hu W, Zienkiewicz J, Chen X, Zhang MZ, Harris RC. et al. EGF receptor-mediated FUS phosphorylation promotes its nuclear translocation and fibrotic signaling. J Cell Biol. 2020 219
36. Wang W, Zhang S, Xu L, Feng Y, Wu X, Zhang M. et al. Involvement of circHIPK3 in the pathogenesis of diabetic cardiomyopathy in mice. Diabetologia. 2021;64:681-92
37. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
38. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
39. Xiao L, Ma XX, Luo J, Chung HK, Kwon MS, Yu TX. et al. Circular RNA CircHIPK3 Promotes Homeostasis of the Intestinal Epithelium by Reducing MicroRNA 29b Function. Gastroenterology. 2021;161:1303-17.e3
40. Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. Journal of molecular cell biology. 2009;1:82-92
41. Brooke GN, Culley RL, Dart DA, Mann DJ, Gaughan L, McCracken SR. et al. FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer Res. 2011;71:914-24
42. Suzuki H, Matsuoka M. Overexpression of nuclear FUS induces neuronal cell death. Neuroscience. 2015;287:113-24
43. Yang Z, Dong X, Pu M, Yang H, Chang W, Ji F. et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2020;23:449-63
44. Tan AY, Riley TR, Coady T, Bussemaker HJ, Manley JL. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci U S A. 2012;109:6030-5
45. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M. et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10:4317
46. Dostert C, Grusdat M, Letellier E, Brenner D. The TNF Family of Ligands and Receptors: Communication Modules in the Immune System and Beyond. Physiological reviews. 2019;99:115-60
47. Punj V, Matta H, Chaudhary PM. X-linked ectodermal dysplasia receptor is downregulated in breast cancer via promoter methylation. Clin Cancer Res. 2010;16:1140-8
48. Tanikawa C, Furukawa Y, Yoshida N, Arakawa H, Nakamura Y, Matsuda K. XEDAR as a putative colorectal tumor suppressor that mediates p53-regulated anoikis pathway. Oncogene. 2009;28:3081-92
49. Sisto M, Lorusso L, Lisi S. X-linked ectodermal dysplasia receptor (XEDAR) gene silencing prevents caspase-3-mediated apoptosis in Sjögren's syndrome. Clin Exp Med. 2017;17:111-9
50. Brennan EP, Mohan M, McClelland A, Tikellis C, Ziemann M, Kaspi A. et al. Lipoxins Regulate the Early Growth Response-1 Network and Reverse Diabetic Kidney Disease. J Am Soc Nephrol. 2018;29:1437-48
51. Watanabe T, Tomioka NH, Doshi M, Watanabe S, Tsuchiya M, Hosoyamada M. Macrophage migration inhibitory factor is a possible candidate for the induction of microalbuminuria in diabetic db/db mice. Biological & pharmaceutical bulletin. 2013;36:741-7
52. Lan X, Kumar V, Jha A, Aslam R, Wang H, Chen K. et al. EDA2R mediates podocyte injury in high glucose milieu. Biochimie. 2020;174:74-83
53. Bowes C, Redd M, Yousfi M, Tauzin M, Murayama E, Herbomel P. Coronin 1A depletion restores the nuclear stability and viability of Aip1/Wdr1-deficient neutrophils. J Cell Biol. 2019;218:3258-71
54. Robichaud S, Fairman G, Vijithakumar V, Mak E, Cook DP, Pelletier AR. et al. Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Autophagy. 2021:1-19
55. Seto S, Tsujimura K, Koide Y. Coronin-1a inhibits autophagosome formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell Microbiol. 2012;14:710-27
Author contact
![]() Corresponding authors: Chun Zhang (E-mail: drzhangchunedu.cn). Address: Department of Nephrology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China; Tel: 86-027-85726006, Fax: 86-027-83617730. Hua Su (E-mail: dr_suhuaedu.cn). Address: Department of Nephrology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China; Tel: 86-027-85726006, Fax: 86-027-83617730
Corresponding authors: Chun Zhang (E-mail: drzhangchunedu.cn). Address: Department of Nephrology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China; Tel: 86-027-85726006, Fax: 86-027-83617730. Hua Su (E-mail: dr_suhuaedu.cn). Address: Department of Nephrology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China; Tel: 86-027-85726006, Fax: 86-027-83617730

 Global reach, higher impact
Global reach, higher impact