10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(15):5885-5896. doi:10.7150/ijbs.78997 This issue Cite
Review
Recent advances in organotypic tissue slice cultures for anticancer drug development
1. Cancer Center, Faculty of Health Sciences, University of Macau, Macau SAR, China.
2. Centre for Precision Medicine Research and Training, Faculty of Health Sciences, University of Macau, Macau SAR, China.
3. MOE Frontier Science Centre for Precision Oncology, University of Macau, Macau SAR, China.
Received 2022-9-16; Accepted 2022-9-19; Published 2022-9-25
Abstract
Organotypic tissue slice culture is established from animal or patient tissues and cultivated in an in vitro ecosystem. This technique has made countless contributions to anticancer drug development due to the vast number of advantages, such as the preservation of the cell repertoire and immune components, identification of invasive ability of tumors, toxicity determination of compounds, quick assessment of therapeutic efficacy, and high predictive performance of drug responses. Importantly, it serves as a reliable tool to stratify therapeutic responders from nonresponders and select the optimal standard-of-care treatment regimens for personalized medicine, which is expected to become a potent platform and even the gold standard for anticancer drug screening of individualization in the near future.
Keywords: organotypic tissue slice culture, anticancer drug discovery, individualized treatment, precision oncology
Introduction
The development of novel drugs is time-consuming, laborious, and costly. In the United States, it requires ~13 years and between 1.8-2.6 billion US dollars from commencement to regulatory approval by the Food and Drug Administration (FDA) [1]. Despite monumental investments, only 5-14% of compounds ultimately manifest therapeutic efficacy plus manageable treatment-related adverse events and are entitled to the permission of clinical administration [1]. The attrition rate of anticancer drug development is even higher; the clinical approval rate of new anticancer drugs is only 3-6% [2], which is attributed to pluralistic reasons, in particular, the usage of suboptimal drug screening and testing platforms. The advent of precision oncology provokes the implementation of individualized medicine into clinical practice. A preluding program of personalized drug screening for cancer patients before treatment by using a reliable drug screening platform to select the optimal treatment regimens theoretically may improve the clinical therapeutic effectiveness.
The most commonly used tool for identifying new antitumor drugs is the two-dimensional (2D) culture of monolayer cells, wherein a standardized high-throughput system is offered to dissect the characteristics of specific cell types [3]. Although contemporary high-throughput screening systems allow the simultaneous assessment of tens or hundreds of thousands of compounds, 2D monolayer cell culture hardly serves as the gold standard platform for antitumor drug discovery in vitro due to its many inherent drawbacks. First, monolayer cell culture rarely recapitulates tumoral heterogeneity and complexity, not to mention the tumor microenvironment (TME) [4]. The TME contains physical, chemical, and biological elements around cancer cells, which are responsible for the cell-cell and cell-extracellular environment interactions that can shape many important biological properties of tumors, e.g., cell differentiation, proliferation, viability, genetic and proteinic expression, suppression and promotion of metastasis, and drug metabolism [5-8]. Second, the genetic and proteinic expression of tumor cells may be changed during the 2D monolayer cell culture [9]. Furthermore, the monolayer morphology interferes with the authentic drug response of cancer cells because there is devoid of multicellular drug resistance [10]. In parallel, the monolayer morphology indicates the unlimited accessibility of oxygen, drugs, and nutrients to cells, which cannot mimic the in vivo phenomenon of gradient-degressive diffusion and perfusion-controlled delivery.
The three-dimentional (3D) organoid technique has been developed rapidly for the identification of anticancer drugs and personalized medicine in the past decade [11, 12]. Organoids are derived from stem cells or progenitor cells and form miniaturized organ-like structures that recreate the chief aspects of the 3D anatomy and multiple cell repertoire of the physiological counterpart and recapitulate basic tissue-level functions and many important characteristics (e.g., mutation spectrum and gene expression) [12-16]. In addition, patient-derived organoids have demonstrated high predictive performance to predict the clinical responses of anticancer drugs in diverse carcinomas [17-20]. However, constructing organoids from tumor biopsies may meet some vexing problems, such as a low number of tumor cellularity and an unmet success rate [10, 14]. This method cannot recapitulate many key cell types in the TME (e.g., immune cells). Another anticancer drug discovery platform, the patient-derived xenograft (PDX), also suffers from several shortcomings, including overlong generation time, high cost, and uncertainty of successful establishment. The successful establishment rate of PDX is contextual and depends on the type and origin of tumors. Nevertheless, it shall be never overlooked the potency of this model in obtaining FDA approval for many drugs.
In 2009, Ootani et al. [21] first introduced a 3D air-liquid interface (3D-ALI) method for an intestinal epithelial culture where the tissue was minced by simple manual scissoring into small pieces (under 0.3 mm3) and then mixed with collagen before cultivation at the air-liquid interface. Because of the success in the recapitulation of a Wnt-dependent stem cell niche, differentiation, and ultrastructure of stomach cell lineages [21, 22], they further employed this technique in the in vitro tumor tissue culture and strikingly found that the immune cells, T-cell receptor repertoire, and phenotypic and genotypic profiling were highly preserved [23]. However, a raised concern is that these undersized minced tissues may not be sufficient to portray the intratumor heterogeneity as it is unclear how many small pieces would be needed to fully recapitulate the heterogeneity of original tumors. Thus, the consistency of drug responses between the 3D-ALI method and the internal condition is required for validation.
The organotypic tissue slice culture platform is also pragmatic for anticancer drug discovery owing to the integration of the alike in vivo microanatomy and the easy manipulation of in vitro work [24-26]. We recently developed a 3D tumor slice culture (3D-TSC) platform that incorporates a lipofuscin autofluorescence feature for the time-course monitoring of drug responses [27]. Compared with other mainstreams, the 3D-TSC platform presents a comparable potential for anticancer drug development (Table 1). This review attempts to summarize and contextualize the contributions of organotypic tissue slice culture in anticancer drug discovery and emphasizes the likely future development in this field.
Overview of the tumor models for anticancer drug discovery
| Models | Cell culture | Organoid | PDX* | 3D-ALI | 3D-TSC |
|---|---|---|---|---|---|
| Successful establishment rate | Low | Moderate | Variable | High | High |
| Generation time | Moderate | Moderate | Long | Short | Short |
| The minimal tumor size requirement | Small | Small | Big | Small | Big |
| TME recapitulation | - | - | -/++ | ++ | ++ |
| Multicellular drug resistance | - | ++ | ++ | + | ++ |
| Intact morphology | - | - | ++ | + | ++ |
| Reproducibility | ++ | ++ | ++ | - | - |
| High-throughput drug screening | ++ | + | - | - | - |
The inapplicability is marked with “-”, the applicability is marked with “+”, and the robust applicability is marked with “++”.
*PDX can recapitulate tumor microenvironment from the humanized mice rather than the severe immunodeficient mice.
Abbreviations: PDX, patient-derived xenograft; 3D-ALI, three-dimentional air-liquid interface method; 3D-TSC, three-dimentional tumor slice culture; TIME, tumor immune microenvironment.
Organotypic tissue slice cultures
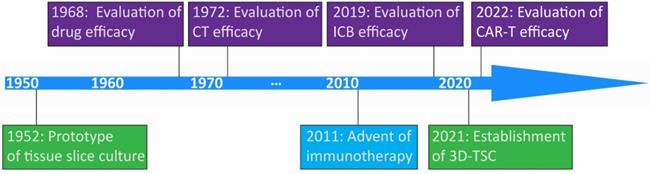
The prototype of organotypic tissue slice culture was developed by Dr. Harford and coworkers in the 1950s [28]. This technique was initially utilized for the assessment of pharmacological efficacy around 1970 [29, 30]. The expediting evolvement of immunotherapy facilitates the application of this technique to evaluate the immune checkpoint blockade, and adoptive cellular therapy response on tumors in recent years [31, 32] (Figure 1). Organotypic tissue slice culture systems represent in vitro cultures of explants of patient- or animal-derived tumoral and normal tissues.
Construction methods
The preparation of tissue slice culture mainly involves the following steps (Figure 2). Briefly, the surgically resected tissues are collected and placed into cold media, which then are meticulously cut into cylindrical or cuboid shapes. These cylindrical or cuboid tissues are sectioned via a slicing method (e.g., vibratome, precision tissue-slicing machine, or simply manual slicing) under sterile conditions within ~6 hours after tumor excision. Only well-shaped slices are selected for cultivation. Slices were incubated in a humidified incubator at 37 °C and 5% CO2 for several days, and the culture media were changed every 2-4 days.
Milestones in discovery and development of organotypic tissue slice culture. Key milestones in the prototype of organotypic tissue slice culture and the establishment of three-dimentional tumor slice culture are shown in light green. In 2011, US FDA approved the first immune checkpoint blockade drug, ipilimumab, in the clinical setting. Key milestones in the evaluation of drug efficacy, chemotherapy efficacy, immune checkpoint blockade efficacy, and chimeric antigen receptor T cell therapy efficacy are shown in purple. Abbreviations: CT, chemotherapy; ICB, immune checkpoint blockade; CAR-T, chimeric antigen receptor T cell; 3D-TSC, three-dimentional tumor slice culture.
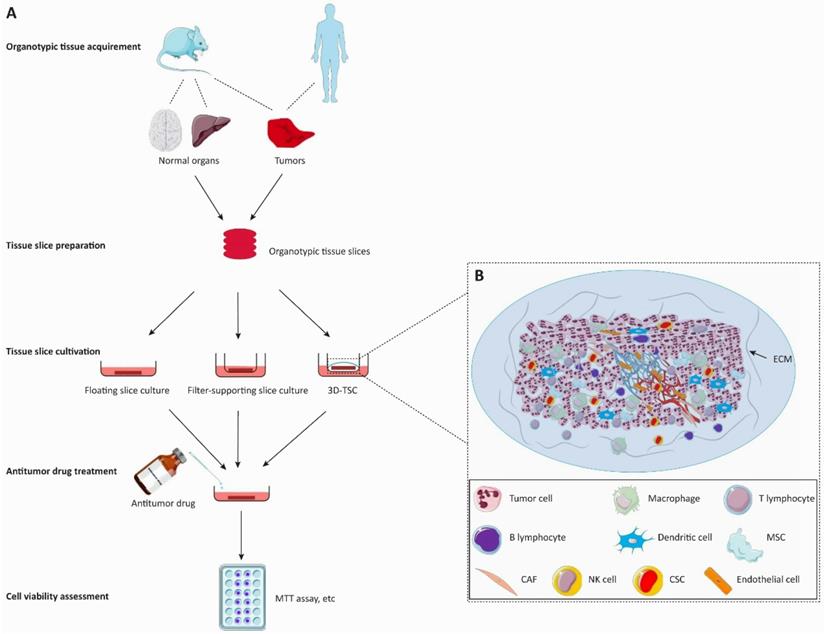
Flow diagram of the organotypic tissue slice culture system for anticancer drug discovery. The organotypic tissue slice culture platform can be used to assess the tumoricidal efficacy of anticancer drugs. A. In the left panel, animal or patient-derived organotypic tissues are cut into the same figurate slices for the antitumor activity assessment of cancer drugs. Tumor slices are employed to evaluate the tumoricidal efficacy of anticancer drugs whereas normal tissue slices are used to observe the invasive ability of tumors and the efficacy of drugs against tumor invasiveness. Drug treatment is immediately initiated after the generation of the organotypic tissue slice culture system. The cell viability of slices is assessed on the 2-7th day of cultivation. B. The 3D-tumor slice culture (3D-TSC) system preserves the architecture and cell repertoire of the original tumor. This platform maximizes the retention of inter-tumor and intratumor heterogeneity, cellular-stromal interactions, and the complexity of the original tumor. The blood vessels on the slices will collapse within a short period after the cessation of blood circulation. Abbreviations: 3D-TSC, three-dimentional tumor slice culture; ECM, extracellular matrix; MSC, Mesenchymal cell; CAF, cancer-associated fibroblast; NK cell, natural killer cell; CSC, cancer stem cell.
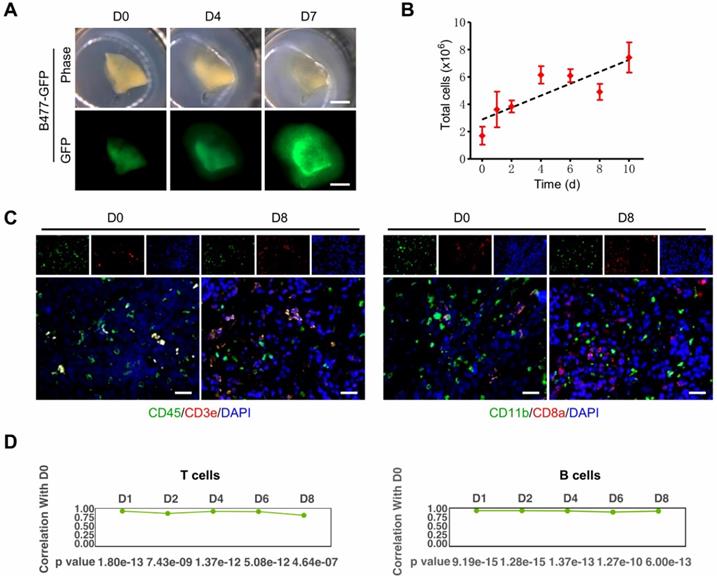
3D-tumor slice culture increases the number of live cells and maintains the immune components of their original tumors. This figure was modified from our previous publication [27] and gained approval from the correspondence authorship. A. Observation of tumor growth in 3D-TSCs derived from primary tumors formed by B477-GFP cells injected into the mammary fat pad of nude mice. Scale bar: 1 mm. B. Time-lapse of cell viability in 3D-TSCs derived from B477-GFP mouse tumors. C. Immunofluorescence staining of immune biomarkers derived from genetically engineered Brca1Co/Co; MMTV-Cre mouse in immunocompetent hosts. Blue is nuclear counterstaining by hematoxylin, and brown staining is positive protein staining by DAB. CD3e/CD8a: T lymphocytes; CD11b, macrophages and microglia marker; CD45, T, NK, dendritic, and lymphokine-activated killer cells marker; Scale bar: 100 µm. D. Pearson correlation coefficient identifying the correlation of gene expression between Day 1 to Day 8 with Day 0 in T cells and B cells.
Underpinning for anticancer drug discovery
Organotypic tissue slice cultures are reliable surrogates for patients or animal models to study some in vivo biological characteristics [33-38]. One tumor bulk can be sliced into tens or even hundreds of slices, enabling the simultaneous assessment of multiple anticancer drugs and the easy realization of 3Rs (i.e., reduction, replacement and refinement) in experiments. Microinjection of tumor cells into normal tissue slices or implantation of 3D tumor cell spheroids on the surface of those slices (hereon called tumor invasive tissue slices) can recapitulate the tumor migration behaviors, contributing to the discovery of effective antitumor drugs against tumor invasion.
The organotypic tissue slice culture platform can be used to validate results obtained from 2D culture and recapitulate in vivo circumstances during cell proliferation and metastasis [39, 40]. It maximally retains many aspects of the original tumors, including inter-tumor and intratumor heterogeneity, TME, morphology, cellular-stromal interplay, and complexity [24, 41-45]. The 3D-TSC platform experiences expanding advantages in these regards [27]. First, the slice kept growing for at least 10 days with a gradually increasing number of total live cells (Figure 3A-B). Second, the key immune cell repertoire and the gene expression level of T lymphocytes and B lymphocytes can be entirely preserved for at least 8 days (Figure 3C-D).
Optimal parameters of the influencing factors
The generation procedures and culture conditions affect the viability of organotypic tissue slices. Slicing tools, thickness, postresection culture timeframe, and oxygen levels all determine the quality and live cell quantity of slice from the beginning of cultivation to the endpoint of drug assessment. Carefully optimizing the preparation and culture conditions can minimize the artificially induced variations in cell viability.
Most studies generate slices with a thickness of 250-500 μm [25-27, 33, 45-47]. Slices with less than 200 μm thickness are extremely fragile and curly and contain large necrotic areas; even at a thickness of 250 μm, a necrosis gradient across the slice has yet been observed [45]. No publication has applied tumor slices with a thickness of more than 500 μm for anticancer drug discovery due to the limited provision of oxygen and nutrients to the center area that can lead to tissue necrosis.
Various postresection culture timeframes give rise to slices with diverse persistence to preserve the tissue architecture and viability, ranging from 4 to 16 days [27, 33-36, 46-49]. Merz et al. [48] sliced glioblastoma specimens and transferred the slices into the culture system within a few minutes after surgical biopsy and, surprisingly, found that the viability and main histological features of the original tumors were conserved for at least 16 days.
Appropriate oxygen levels are necessary for the retention of slice morphology [26], as hypoxia (< 5% O2) elicits rapid and significant changes to many stress pathways and is associated with a negative impact on the morphology of tumor slices [26]. Moreover, hyperoxic conditions (41% O2) do not increase the tissue viability, metabolic activity, and proliferation of slices compared to normoxic conditions (21% O2) [25]. Therefore, the atmospheric oxygen level is sufficient for slice culture. Intermittent exposure of slices to oxygen and nutrients using a rotating incubation unit further potentiates slice viability compared to the floating or stagnant filter-supporting slice culture [45, 47]. One explanation is that the rotating incubation units result in a higher elimination efficiency of waste products. Floating slices in the liquid culture medium essentially cannot meet the oxygen supplement because of the limited solubility of oxygen (< 2.2 mmol/L) [50]. Positioning the slice on a filter at the air-liquid interface achieves the efficient uptake of oxygen and nutrients [27, 33]. Koch et al. [50] floated a liver slice on perfluorodecalin, a non-water-soluble chemical of the artificial oxygen carrier, and overlaid it with the culture medium also conquers the predicament of insufficient oxygen supplementation in the culture medium and prolongs the lifespan of the slice as the perfluorodecalin can continuously provide oxygen to the slice from the bottom.
The optimal time window for the assessment of therapeutic efficacy needs to be clarified, which should obviate the slicing-caused activation of stress and inflammatory responses in the beginning and the tissue destruction and devitalization of overlong cultivation. Metabolic activity is continuously elevated within the first 24 hours of cultivation but does not fluctuate at later time points [25]. Moreover, slicing-caused tissue damage mainly occurs at the outer part of the slices within the first 24 hours but thereafter is not observed during the subsequent 72 hours of follow-up [25]. These findings suggest that the first 24 hours is the contraindicated period for the assessment of therapeutic responses. Also, consistency, high-throughput screening, intratumoral heterogeneity, antibody delivery, cryopreservation, and live tissue imaging are some of the challenges of this system that should be addressed by a balanced review but not the purpose of this work.
Promising applications
The organotypic tissue slice platform allows the selection of optimal treatment regimens for individualization in the context of precision oncology and has won popularity in the elucidation of tumor invasive ability, the potential toxicity of drugs, and the efficacy assessment of multiple treatment paradigms.
Identification of tumor invasiveness
The tumor invasive tissue slices visualize the tumor invasiveness in near real-time and offer easy maneuverability for testing the pharmacological efficacy of therapies against tumor migration [51-54]. Tumor invasiveness varies with the different morphologies of tumor cells, in which the invasive phenotype cells are characterized by a small soma with a distinct leading process [55]. The intratumoral composition of the invasive phenotype cells and the signaling between them and the tumor core are the determinants conditioning tumor invasiveness [55]. Therefore, finding a treatment that targets the invasive phenotype cells may significantly mitigate tumor migration.
A profound marker (i.e., “generalized stiffness”) of the invasive phenotype tumor cells has been found on the tumor invasive normal tissue slice culture system [56, 57]. Anticancer drug-caused alterations of the tumor cell mechanical and migratory patterns in this model are the theoretical supports for its clinical application in testing the therapeutic efficacy of drugs inhibiting tumor invasion. The mechanical and migratory properties of glioblastoma (GBM) cells on hippocampal slices can be weakened by cannabinoids [57]. Similarly, 2 Gy of nonlethal irradiation on this platform attenuates GBM cell invasiveness by increasing the “generalized stiffness” and inducing changes in the actin cytoskeleton and the motility of tumor cells [56]. Implanting C6 GBM cells on the surface of brain slices manifests the antitumor invasiveness activity of the Rac1 inhibitor NSC23766) (Table 2) [58]. The tumor invasive normal tissue slice culture system with these unique advantages will account for more room in the preclinical stage of unveiling many effective antitumor invasive drugs.
Toxicity determination
Plus, the tumor invasive tissue slices show a promising outlook in the evaluation of the potential pharmacological toxicity. Brain slices can serve as a tool to reflect the toxicity of various neurotoxic compounds and may provide interventions to mitigate toxicity [59, 60]. A great predictive performance in the prediction of drug-induced seizure liability is also observed in this model, with 93% sensitivity and 100% specificity [61]. The organotypic hippocampal slices have underpinned several mechanistic insights underlying ethanol dependence and/or ethanol withdrawal-induced neurochemical toxicity, such as abnormal synaptic transmission and CA1 pyramidal cell death [62, 63]. Drug-induced cholestasis can be modeled on liver slices via 48 hours of incubation with human cholestasis-causing drugs, allowing the observation of the pathogenesis and progression of the disease under an in vitro condition [64]. Doxorubicin and trastuzumab both show cardiotoxicity in the clinical treatment of breast cancer patients, and this phenomenon is also recapitulated on the heart slice culture model, as manifested by a loss of cardiomyocyte structure and function after 48 hours of treatment [65].
Assessment of therapeutic efficacy
Organotypic tumor slice culture demonstrates the suitability to investigate the antitumor activity of small-molecule drug therapy (i.e., chemotherapy and molecularly targeted therapy) [24, 25, 27, 41, 47, 49, 66-84], immunotherapy [27, 31, 85], radiotherapy [86-88], and adoptive cellular therapy [32], as summarized in Table 3 and discussed below.
Small-molecule drug therapy
Organotypic slices treated with small-molecule drugs show concentration-dependent and heterogeneous responses [77, 86], exemplified by reduction of cell viability [79], decrease in cell proliferation [67, 80-82], and increase in cell loss and apoptosis [67, 80-82]. Tumor slice culture has been incorporated into the discovery of new combination therapy regimens and the efficacy assessment of clinical standard-of-care treatment for multiple cancers [47, 66-68]. For example, the oncological efficacy and safety profiles of the CAF regimen (i.e., cyclophosphamide, adriamycin plus 5-fluorouracil) in breast cancers have been confirmed by many landmark phase 2 to 3 randomized controlled trials [89-93], and therefore this regimen is recommended for clinical application by the National Comprehensive Cancer Network (NCCN) Guidelines version 2. 2022 (https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf). Utilizing breast cancer patient-derived slices to assess the CAF response succeeds in the stratification of CAF responders from nonresponders [47]. Despite the tumor slices are ubiquitously suitable to assess small-molecule drug responses, it is more intriguing to characterize the immunotherapeutic efficacy.
Contributions of organotypic tissue slice model from normal organs in antitumor invasive drug discovery
| Models | Sketch diagram | Contributions | Reference |
|---|---|---|---|
| Organotypic brain slice culture | 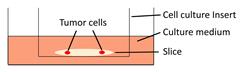 | Jasplakinolide, Rac1 inhibitor NSC23766, and tranilast significantly decrease the tumor cell invasion on brain slices. | [52, 58, 77] |
| Organotypic cerebellar slice culture | 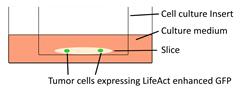 | Epidermal growth factor accelerates the invasion of medulloblastoma cells on cerebellar slices. | [53] |
| Tissue-based liver-kidney-on-a-chip | 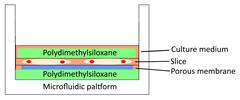 | A CXCR4 small-molecule antagonist AMD3100 effectively halts the liver tropism of breast cancer extracellular vesicles. | [54] |
| Organotypic hippocampal slice culture | 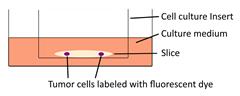 | Cannabinoids influence the migratory and mechanical properties of tumor cells on organotypic hippocampal slices. | [57] |
The in vitro response of treatments on the patient-derived tumor slice culture system
| Type of cancer | Treatment* | No. of patients | In vitro response | Reference |
|---|---|---|---|---|
| Prostate cancer and bladder cancer | Docetaxel or gemcitabine | 10 | Induction of cell death and increase in cell loss | [24] |
| Pancreatic ductal adenocarcinoma | Rapamycin | 12 | Decrease in metabolic activity | [25] |
| Colon cancer and breast cancer | Chemotherapy, endocrinotherapy, targeted therapy, immunotherapy, and polytherapy† | 7‡ | Decrease in cell viability and increase in apoptosis, with a heterogenous individual response to chemotherapy or immunotherapy. | [27] |
| Colorectal cancer liver metastasis | IL-10 antibody plus CAR-T cell therapy | 38 | αIL-10 augments CAR-T cell activation and CAR-T cell-mediated cytotoxicity | [32] |
| Hepatic metastatic colorectal carcinoma | Oxaliplatin, cetuximab, or pembrolizumab | 9 | Decrease in cell proliferation, with a heterogenous individual response to chemotherapy and targeted therapy | [41] |
| Breast cancer | Cyclophosphamide, adriamycin plus 5-FU | 15 | Decrease in cell proliferation and induction of cell death | [47] |
| Glioblastoma | Temozolomide | 12 | Decrease in cell proliferation and increase in cell loss and apoptosis, with a heterogenous individual response to chemotherapy | [48] |
| Gastric and esophagogastric junction cancer | 5-FU or cisplatin | 13 | Increase in cell loss and apoptosis | [49] |
| Hepatocellular carcinoma | Sorafenib plus N20 blocking peptide | 13 | Decrease in cell proliferation | [66] |
| Colorectal carcinoma | 5-FU | 7 | A dose-dependent decrease in cell proliferation, with a heterogenous individual response to chemotherapy | [67] |
| Bladder cancer | Mitomycin-C plus coxsackie A21 | 1 | Stronger apoptosis in the combination therapy than either of the monotherapy | [68] |
| HNSCC | Cetuximab | 10 | Decrease in cell proliferation, with a heterogenous individual response to targeted therapy | [69] |
| HNSCC | Cetuximab | 14 | Decrease in cell proliferation, with a heterogenous individual response to targeted therapy | [70] |
| Glioblastoma | Gefitinib | 1 | Insensitive anticancer activity | [71] |
| Melanoma | Ribociclib plus CGM097 | 13 | The impedance of cell growth | [72] |
| Prostate cancer | Enzalutamide, or olaparib | 3 | Decrease in cell proliferation and increase in cell loss, with a heterogenous individual response to anti-androgen or targeted therapy | [73] |
| Breast cancer | Rapamycin | 30 | Decrease in cell proliferation, with a heterogenous individual response to targeted therapy | [75] |
| Rectal cancer liver metastasis | Oxaliplatin | 20 | Decrease in tumor size and cell viability, and increase in apoptosis | [78] |
| Breast cancer | Doxorubicin | 1 | A dose-dependent decrease in cell viability | [79] |
| Pancreatic ductal adenocarcinoma | Staurosporine, gemcitabine or cisplatin | 10 | Decrease in cell proliferation and increase in cell loss and apoptosis | [80] |
| Pancreatic ductal adenocarcinoma | Staurosporine or cycloheximide | 13 | A dose- and time-dependent increase in apoptosis and decrease in cell proliferation | [81, 82] |
| Lung cancer | Cisplatin | 32 | Induction of cell death | [83] |
| Melanoma, NSCLC, RCC, breast cancer, and ovarian cancer | Nivolumab | 37‡ | Increase in immune activity, with a heterogenous individual response to immunotherapy | [85] |
| Oral squamous cell carcinoma | 4 Gy irradiation | 28 | More cancer stem cells and DNA damage response in responders than nonresponders | [87] |
*The “or”-connected drugs represent monotherapy, while the “plus”-connected drugs represent combination therapy.
‡These 7 patients consist of 2 breast cancer patients and 5 colon cancer patients, and these 37 patients consist of 13 melanoma patients, 7 NSLCC patients, 8 breast cancer patients, 6 ovarian patients, and 3 RCC patients.
†The drugs involved in these treatments include 5-fluorouracil, cisplatin, docetaxel, doxorubicin, epirubicin, mitoxantrone, irinotecan, daunorubicin, tamoxifen, neratinib, ceritinib, afatinib, regorafenib, osimertinib, palbociclib, pembrolizumab, durvalumab, and durvalumab plus IL-2.
Abbreviations: 5-FU, 5-fluorouracil; HNSCC, head and neck squamous cell carcinoma; RCC, renal cell carcinoma.
Immunotherapy
Deciphering immune-oncology on organotypic tissue slice was initially described by Sivakumar et al. [31], who demonstrated that the live immune cell populations lasted for 7 days on the filter-supporting tumor slice cultures. Although some immune cell populations are changed between the slice model and the original tumor and the proportion of the immune components are variable with the time-lapse, this technique observed the response of immune checkpoint blockade. Introducing novel biological materials and techniques into the tumor slice culture system may avoid the variation of immune cell populations and key immune mediators, contributing to the more confidential in vitro results of immunotherapeutic efficacy. Indeed, Voabil and colleagues [85] embedded patient-derived tumor fragments into an artificial extracellular matrix and found that the lymphocyte efflux was significantly lower than those cultured in medium or collagen, with no significant difference in T-cell functionality and nonspecific immune activation. They evaluated the programmed cell death protein 1 (PD-1) blockade response of 37 tumors from five cancer types (i.e., melanoma, non-small-cell lung cancer, breast cancer, ovarian cancer, and renal cell carcinoma) on this platform. Only a small proportion of tumors (13 out of 37) showed discernable immunotherapy responses, predominantly from melanoma and non-small-cell lung cancer, which was concordant with the clinical outcomes.
The filter-supporting tumor slice culture model with a 0.4 μm pore size membrane culture insert cannot prevent the horizontal lymphocyte efflux and thus may identify the negative result of pembrolizumab (a humanized anti-PD-1 monoclonal antibody) responses [41]. Tumor slice in the 3D-TSC platform is precoated with a reconstituted collagen solution and supported by an insert at the air-liquid interface; therefore, the horizontal and vertical lymphocyte efflux is efficiently spared and the lymphocytes are well preserved within 8 days of cultivation [31]. We utilized this technique to evaluate the immunotherapeutic efficacy of colon cancer or breast cancer patients. The expression level of programmed cell death ligand 1 (PD-L1) in colon cancer patients was low, only between 2-11%. Patients with 11% PD-L1 expression showed the highest PD-1/PD-L1 blockade response; intriguingly, the anti-PD1/PD-L1 therapeutic efficacy was higher in patients with 2% PD-L1 expression than in those with 5% or 7% PD-L1 expression. One explanation is that intratumoral PD-L1 is heavily glycosylated, and the immunohistochemical readout results may not reflect the reality of PD-L1 expression [94]. Removal of N-linked glycosylation significantly elevates PD-L1 detection in human tumor tissues, and the improved PD-L1 detection level is closely related to the clinical oncological efficacy of anti-PD-1/PD-L1 therapy [94]. Therefore, PD-L1 after deglycosylation can be used as a biomarker to predict immunotherapeutic efficacy.
Radiotherapy
Radiotherapy reduces the risk of tumor relapse, ameliorates survival benefits, and has become an irreplaceable component of systematic treatment for many cancers [95]. Organotypic tissue slices exposed to irradiation observe the stall of cell growth and the decrease in tumor volume [86]. Cancer stem cell status and DNA damage repair both dictate tumor radioresistance. After 24 hours of exposure to 4 Gy irradiation, the expression levels of the cancer stem cell marker CD44 and DNA damage repair markers γ-H2AX and p-ATM in tumor slices from radiotherapy-sensitive oral squamous cell carcinoma patients are significantly higher than those from radiotherapy-resistant patients [87], suggesting that this platform is applicable to stratify radiotherapeutic responders from nonresponders. Radiotherapy plus the inhibitor of cyclin-dependent kinases 4 and 6 (CDK4/6i) palbociclib contribute to a significantly increased cell death in meningioma slices than either of the monotherapy [88]. These results offer preclinical evidence for the cotreatment of CDK4/6i with radiotherapy for meningiomas.
Adoptive cellular therapy
Adoptive cellular therapy is a new type of therapy that aims to leverage the internal immune system to obviate cancers, wherein immune cells are engineered with the expression of anti-specific T-cell receptors or chimeric antigen receptors (CARs) that can better recognize and kill malignant cells [96, 97]. The overwhelming triumph of CD19-targeted CAR-T cell therapy in refractory B-cell malignant lymphoma is an important landmark of adoptive cellular therapy in cancers [98, 99] and serves as an exemplification for developing more treatment paradigms, e.g., CAR-NK cell therapy [96]. The tumor slice culture platform recently has been used to assess the efficacy of CAR-T cell therapy in solid tumors. Despite the low success of CAR-T cell therapy in human colorectal cancer liver metastases (CRLM), the interleukin-10 blockade has shown functions to enhance the antitumor activity of CAR-T cell therapy in human CRLM slices, as characterized by the increased CAR-T cell activation and CAR-T cell-caused cytotoxicity as well as amplified CAR-T cell proliferation [32]. Therefore, harnessing tumor slice culture models may broaden the clinical application of adoptive cellular therapy in solid tumors.
Conclusions and future perspectives
Organotypic tissue slice culture platform has many advantages for anticancer drug discovery, including (1) identification of tumoricidal efficacy of anticancer drugs, specifically the immune checkpoint blockade antibodies; (2) high predictive performance of drug responses; (3) quick stratification of treatment responders and nonresponders; and (4) ascertainment of the cooperativity of different combination therapy strategies. Therefore, this technique may tailor the best clinical guideline-recommended regimens for individualization, providing a decision aid for clinicians. Recapitulation of the complexity and TME of original tumors on the slices is of pivotal importance, particularly the indispensable components for specific anticancer drugs (e.g., immune cell repertoire for PD-1/PD-L1 blockade). Thus, it needs to consider all aspects of the generation and cultivation conditions for maximally maintaining the consistency of the slices with the original tissues. Notably, accurately judging therapeutic efficacy requires averaging an adequate number of slice results due to intratumoral heterogeneity. In addition, the system is not a reproducible tool similar to 3D-ALI or patient-derived xenografts and is normally unable to test a plethora of drug responses. It is crucial to enhance the efficiency of clinical translation of preclinical findings on this technique, for which it needs to narrow down the drug candidates by prescreening compounds by the 2D monolayer cell culture or selecting agents according to the NCCN clinical practice guidelines in oncology.
The promising applications of different organotypic tumor slice cultures in antitumor drug discovery
| Models | Sketch diagram | Small-molecule drug therapy | Immunotherapy | Radiotherapy | Adoptive cellular therapy | Reference |
|---|---|---|---|---|---|---|
| Filter-supporting tumor slice culture | 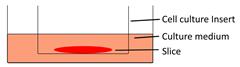 | √ | √ | √ | √ | [25, 35, 41, 49, 66, 81] |
| Floating tumor slice culture | 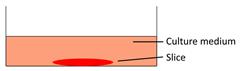 | √ | √ | √ | × | [26, 33, 68, 69, 72, 75] |
| 3D-tumor slice culture | 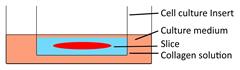 | √ | √ | × | × | [27] |
| Tumor slice culture on a rotating platform | 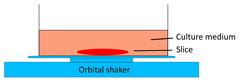 | √ | × | × | × | [47] |
| Collagen-supporting tumor slice culture | 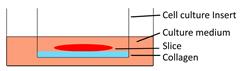 | √ | × | × | × | [80] |
| Tumor slice culture on a microfluidic platform | 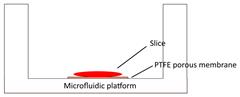 | √ | × | × | × | [84] |
| Patient-derived tumor fragment culture | 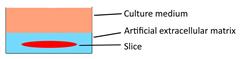 | × | √ | × | × | [85] |
The application that has been described in the publications is marked with “√”, otherwise is marked with “×”.
Different tissue origins and preparation procedures bestow the platform with the intrinsic advantages and limitations in the field of anticancer drug discovery. We enumerated the overlapping but distinct preparation conditions for different organotypic tumor slice models in Table 4. Each of them has been documented with the potential to assess the oncological efficacy of the antitumor treatment strategy. In addition, tissue slices have implicated the availability of uncovering novel biomaterials within the synthesis of anticancer drugs; for example, graphene quantum dots and nanoparticles both show complete penetration and minimal viability damage on slices [100, 101], indicating their promising application for the construction of drug delivery systems.
Abbreviations
2D: two-dimensional; TME: tumor microenvironment; 3D: three-dimentional; PDX: patient-derived xenograft; 3D-ALI: 3D air-liquid interface; 3D-TSC: 3D tumor slice culture; GBM: glioblastoma; CAF: cyclophosphamide, adriamycin plus 5-fluorouracil; NCCN: National Comprehensive Cancer Network; PD-1: programmed cell death protein 1; PD-L1: programmed cell death ligand 1; CDK4/6i: inhibitor of cyclin-dependent kinases 4 and 6; CARs: chimeric antigen receptors; CRLM: colorectal cancer liver metastases.
Acknowledgements
We thank the critical reading and helpful discussion by Kai Miao and Heng Sun in Deng Lab (Faculty of Health Sciences, University of Macau). We are grateful for Fuqiang Xing for the assistance in the figure preparation. This work was supported by the Chair Professor Grant (CPG 2021-00021-FHS), Multi-Year Research Grant (MYRG 2017-00113-FHS, MYRG 2018-00070-FHS, and MYRG2019-00067-FHS) from University of Macau; the Science and Technology Development Fund, Macau SAR (094/2015/A3, 122/2016/A3, 018/2017A1, 0011/2019/AKP, 0112/2019/A2, 0034/2019/AGJ, and 048/2019/A1), Shenzhen Science and Technology Innovation Committee EF040/FHS-DCX/2021/SZSTIC, and Natural Science Foundation of China (32070681, and 82030094).
Author Contributions
LH wrote the manuscript and drew Tables and Figures. CXD was responsible for the study design, and supervision. All authors reviewed and approved the manuscript prior to submission.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schomberg DT, Tellez A, Meudt JJ, Brady DA, Dillon KN, Arowolo FK, Wicks J, Rousselle SD, Shanmuganayagam D. Miniature Swine for Preclinical Modeling of Complexities of Human Disease for Translational Scientific Discovery and Accelerated Development of Therapies and Medical Devices. Toxicol Pathol. 2016;44:299-314
2. Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics. 2019;20:273-286
3. Carter EP, Gopsill JA, Gomm JJ, Jones JL, Grose RP. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer. Breast Cancer Res. 2017;19:50
4. Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249-289
5. Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839-845
6. Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, van der Kuip H. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J. 2014;9:1115-1128
7. Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015-3024
8. Guo S, Deng CX. Effect of Stromal Cells in Tumor Microenvironment on Metastasis Initiation. Int J Biol Sci. 2018;14:2083-2093
9. Agarwal P, Wang H, Sun M, Xu J, Zhao S, Liu Z, Gooch KJ, Zhao Y, Lu X, He X. Microfluidics Enabled Bottom-Up Engineering of 3D Vascularized Tumor for Drug Discovery. ACS Nano. 2017;11:6691-6702
10. Kondo J, Inoue M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells. 2019;8:470
11. Chen P, Zhang X, Ding R, Yang L, Lyu X, Zeng J, Lei JH, Wang L, Bi J, Shao N. et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv Sci (Weinh). 2021;8:e2101176
12. Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671-687
13. Duarte AA, Gogola E, Sachs N. BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat Methods. 2018;15:134-140
14. Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M, Gao D. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun. 2018;9:2404
15. Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H. et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25:838-849
16. Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952-955
17. Vlachogiannis G, Hedayat S. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920-926
18. Yin S, Xi R. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci Transl Med. 2020 12
19. Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, Xia F, Fu G, Deng Y, Pan M. et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell. 2020;26:17-26.e16
20. Ooft SN, Weeber F. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019 11
21. Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701-706
22. Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, Cantrell MA, Rack PG, Neal JT, Chan CW. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20:769-777
23. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR. et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175:1972-1988.e1916
24. van de Merbel AF, van der Horst G, van der Mark MH, van Uhm JIM, van Gennep EJ, Kloen P, Beimers L, Pelger RCM, van der Pluijm G. An ex vivo Tissue Culture Model for the Assessment of Individualized Drug Responses in Prostate and Bladder Cancer. Front Oncol. 2018;8:400
25. Misra S, Moro CF, Del Chiaro M, Pouso S, Sebestyén A, Löhr M, Björnstedt M, Verbeke CS. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:2133
26. Davies EJ, Dong M, Gutekunst M, Närhi K, van Zoggel HJ, Blom S, Nagaraj A, Metsalu T, Oswald E, Erkens-Schulze S. et al. Capturing complex tumour biology in vitro: histological and molecular characterisation of precision cut slices. Sci Rep. 2015;5:17187
27. Xing F, Liu YC, Huang S, Lyu X, Su SM, Chan UI, Wu PC, Yan Y, Ai N, Li J. et al. Accelerating precision anti-cancer therapy by time-lapse and label-free 3D tumor slice culture platform. Theranostics. 2021;11:9415-9430
28. Harford CG, Hamlin A. Effect of influenza virus on cilia and epithelial cells in the bronchi of mice. J Exp Med. 1952;95:173-190
29. Field JB, Remer A, Bloom G, Kriss JP. In vitro stimulation by long-acting thyroid stimulator of thyroid glucose oxidation and 32P incorporation into phospholipids. J Clin Invest. 1968;47:1553-1560
30. Willoughby HW, Maughan GB, Tremblay PC, Wood N. Determination of individual human tumour sensitivity to antitumour agents by tissue-slice incubation. Can J Surg. 1971;14:406-409
31. Sivakumar R, Chan M, Shin JS, Nishida-Aoki N, Kenerson HL, Elemento O, Beltran H, Yeung R, Gujral TS. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. Oncoimmunology. 2019;8:e1670019
32. Sullivan KM, Jiang X, Guha P, Lausted C, Carter JA, Hsu C, Labadie KP, Kohli K, Kenerson HL, Daniel SK. et al. Blockade of interleukin 10 potentiates antitumour immune function in human colorectal cancer liver metastases. Gut. 2022
33. Salas A, López J, Reyes R, Évora C, de Oca FM, Báez D, Delgado A, Almeida TA. Organotypic culture as a research and preclinical model to study uterine leiomyomas. Sci Rep. 2020;10:5212
34. Riley A, Green V, Cheah R, McKenzie G, Karsai L, England J, Greenman J. A novel microfluidic device capable of maintaining functional thyroid carcinoma specimens ex vivo provides a new drug screening platform. BMC Cancer. 2019;19:259
35. Ghaderi M, Fernández Moro C, Pouso Elduayen S, Hultin E, Verbeke CS, Björnstedt M, Dillner J. Genome-wide transcriptome profiling of ex-vivo precision-cut slices from human pancreatic ductal adenocarcinoma. Sci Rep. 2020;10:9070
36. Maund SL, Nolley R, Peehl DM. Optimization and comprehensive characterization of a faithful tissue culture model of the benign and malignant human prostate. Lab Invest. 2014;94:208-221
37. Figiel S, Pasqualin C, Bery F, Maupoil V, Vandier C, Potier-Cartereau M, Domingo I, Guibon R, Bruyere F, Maheo K, Fromont G. Functional Organotypic Cultures of Prostate Tissues: A Relevant Preclinical Model that Preserves Hypoxia Sensitivity and Calcium Signaling. Am J Pathol. 2019;189:1268-1275
38. Keshari KR, Sriram R, Van Criekinge M, Wilson DM, Wang ZJ, Vigneron DB, Peehl DM, Kurhanewicz J. Metabolic reprogramming and validation of hyperpolarized 13C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate. 2013;73:1171-1181
39. Spennati G, Horowitz LF, McGarry DJ, Rudzka DA, Armstrong G, Olson MF, Folch A, Yin H. Organotypic platform for studying cancer cell metastasis. Exp Cell Res. 2021;401:112527
40. Centritto F, Paroni G, Bolis M, Garattini SK, Kurosaki M, Barzago MM, Zanetti A, Fisher JN, Scott MF, Pattini L. et al. Cellular and molecular determinants of all-trans retinoic acid sensitivity in breast cancer: Luminal phenotype and RARα expression. EMBO Mol Med. 2015;7:950-972
41. Martin SZ, Wagner DC, Hörner N, Horst D, Lang H, Tagscherer KE, Roth W. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer. BMC Cancer. 2019;19:1030
42. Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10:483-487
43. Annels NE, Arif M, Simpson GR, Denyer M, Moller-Levet C, Mansfield D, Butler R, Shafren D, Au G, Knowles M. et al. Oncolytic Immunotherapy for Bladder Cancer Using Coxsackie A21 Virus. Mol Ther Oncolytics. 2018;9:1-12
44. Russell S, Wojtkowiak J, Neilson A, Gillies RJ. Metabolic Profiling of healthy and cancerous tissues in 2D and 3D. Sci Rep. 2017;7:15285
45. Nagaraj AS, Bao J, Hemmes A, Machado M, Närhi K, Verschuren EW. Establishment and Analysis of Tumor Slice Explants As a Prerequisite for Diagnostic Testing. J Vis Exp. 2018
46. Köcher S, Beyer B, Lange T, Nordquist L, Volquardsen J, Burdak-Rothkamm S, Schlomm T, Petersen C, Rothkamm K, Mansour WY. A functional ex vivo assay to detect PARP1-EJ repair and radiosensitization by PARP-inhibitor in prostate cancer. Int J Cancer. 2019;144:1685-1696
47. Naipal KA, Verkaik NS, Sánchez H, van Deurzen CH, den Bakker MA, Hoeijmakers JH, Kanaar R, Vreeswijk MP, Jager A, van Gent DC. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer. 2016;16:78
48. Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, Gutenberg A, Giese A, Schopow K, Hellwig C, Schäfer M. et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol. 2013;15:670-681
49. Koerfer J, Kallendrusch S, Merz F, Wittekind C, Kubick C, Kassahun WT, Schumacher G, Moebius C, Gaßler N, Schopow N. et al. Organotypic slice cultures of human gastric and esophagogastric junction cancer. Cancer Med. 2016;5:1444-1453
50. Koch A, Saran S, Tran DD, Klebba-Färber S, Thiesler H, Sewald K, Schindler S, Braun A, Klopfleisch R, Tamura T. Murine precision-cut liver slices (PCLS): a new tool for studying tumor microenvironments and cell signaling ex vivo. Cell Commun Signal. 2014;12:73
51. Chadwick EJ, Yang DP, Filbin MG, Mazzola E, Sun Y, Behar O, Pazyra-Murphy MF, Goumnerova L, Ligon KL, Stiles CD, Segal RA. A Brain Tumor/Organotypic Slice Co-culture System for Studying Tumor Microenvironment and Targeted Drug Therapies. J Vis Exp 2015:e53304.
52. Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P. An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer. 2018;18:103
53. Neve A, Santhana Kumar K, Tripolitsioti D, Grotzer MA, Baumgartner M. Investigation of brain tissue infiltration by medulloblastoma cells in an ex vivo model. Sci Rep. 2017;7:5297
54. Tian H, Pang J, Qin K, Yuan W, Kong J, Ma H, He J, Yang X, Luo Y, Lu Y. et al. A Novel Tissue-Based Liver-Kidney-on-a-Chip Can Mimic Liver Tropism of Extracellular Vesicles Derived from Breast Cancer Cells. Biotechnol J. 2020;15:e1900107
55. Fayzullin A, Tuvnes FA, Skjellegrind HK, Behnan J, Mughal AA, Langmoen IA, Vik-Mo EO. Time-lapse phenotyping of invasive glioma cells ex vivo reveals subtype-specific movement patterns guided by tumor core signaling. Exp Cell Res. 2016;349:199-213
56. Hohmann T, Grabiec U, Vogel C, Ghadban C, Ensminger S, Bache M, Vordermark D, Dehghani F. The Impact of Non-Lethal Single-Dose Radiation on Tumor Invasion and Cytoskeletal Properties. Int J Mol Sci. 2017;18:2001
57. Hohmann T, Grabiec U, Ghadban C, Feese K, Dehghani F. The influence of biomechanical properties and cannabinoids on tumor invasion. Cell Adh Migr. 2017;11:54-67
58. Ren B, Yu S, Chen C, Wang L, Liu Z, Wu Q, Wang L, Zhao K, Yang X. Invasion and anti-invasion research of glioma cells in an improved model of organotypic brain slice culture. Tumori. 2015;101:390-397
59. Choudhury ME, Sugimoto K, Kubo M, Iwaki H, Tsujii T, Kyaw WT, Nishikawa N, Nagai M, Tanaka J, Nomoto M. Zonisamide up-regulated the mRNAs encoding astrocytic anti-oxidative and neurotrophic factors. Eur J Pharmacol. 2012;689:72-80
60. Farizatto KLG, McEwan SA, Naidoo V, Nikas SP, Shukla VG, Almeida MF, Byrd A, Romine H, Karanian DA, Makriyannis A, Bahr BA. Inhibitor of Endocannabinoid Deactivation Protects Against In vitro and In vivo Neurotoxic Effects of Paraoxon. J Mol Neurosci. 2017;63:115-122
61. Zhai J, Zhou YY, Lagrutta A. Sensitivity, specificity and limitation of in vitro hippocampal slice and neuron-based assays for assessment of drug-induced seizure liability. Toxicol Appl Pharmacol. 2021;430:115725
62. Gerace E, Landucci E, Bani D, Moroni F, Mannaioni G, Pellegrini-Giampietro DE. Glutamate Receptor-Mediated Neurotoxicity in a Model of Ethanol Dependence and Withdrawal in Rat Organotypic Hippocampal Slice Cultures. Front Neurosci. 2018;12:1053
63. Gerace E, Landucci E, Totti A, Bani D, Guasti D, Baronti R, Moroni F, Mannaioni G, Pellegrini-Giampietro DE. Ethanol Toxicity During Brain Development: Alterations of Excitatory Synaptic Transmission in Immature Organotypic Hippocampal Slice Cultures. Alcohol Clin Exp Res. 2016;40:706-716
64. Karsten REH, Krijnen NJW, Maho W, Permentier H, Verpoorte E, Olinga P. Mouse precision-cut liver slices as an ex vivo model to study drug-induced cholestasis. Arch Toxicol. 2022;96:2523-2543
65. Miller JM, Meki MH, Ou Q, George SA, Gams A, Abouleisa RRE, Tang XL, Ahern BM, Giridharan GA, El-Baz A. et al. Heart slice culture system reliably demonstrates clinical drug-related cardiotoxicity. Toxicol Appl Pharmacol. 2020;406:115213
66. Karkampouna S, van der Helm D, Gray PC, Chen L, Klima I, Grosjean J, Burgmans MC, Farina-Sarasqueta A, Snaar-Jagalska EB, Stroka DM. et al. CRIPTO promotes an aggressive tumour phenotype and resistance to treatment in hepatocellular carcinoma. J Pathol. 2018;245:297-310
67. Sönnichsen R, Hennig L, Blaschke V, Winter K, Körfer J, Hähnel S, Monecke A, Wittekind C, Jansen-Winkeln B, Thieme R. et al. Individual Susceptibility Analysis Using Patient-derived Slice Cultures of Colorectal Carcinoma. Clin Colorectal Cancer. 2018;17:e189-e199
68. Relph K, Annels N, Smith C, Kostalas M, Pandha H. Oncolytic Immunotherapy for Bladder Cancer Using Coxsackie A21 Virus: Using a Bladder Tumor Precision-Cut Slice Model System to Assess Viral Efficacy. Methods Mol Biol. 2020;2058:249-259
69. Peria M, Donnadieu J, Racz C, Ikoli JF, Galmiche A, Chauffert B, Page C. Evaluation of individual sensitivity of head and neck squamous cell carcinoma to cetuximab by short-term culture of tumor slices. Head Neck. 2016;38(Suppl 1):E911-915
70. Donnadieu J, Lachaier E, Peria M, Saidak Z, Dakpe S, Ikoli JF, Chauffert B, Page C, Galmiche A. Short-term culture of tumour slices reveals the heterogeneous sensitivity of human head and neck squamous cell carcinoma to targeted therapies. BMC Cancer. 2016;16:273
71. Loriguet L, Morisse MC, Dremaux J, Collet L, Attencourt C, Coutte A, Boone M, Sevestre H, Galmiche A, Gubler B. et al. Combining genomic analyses with tumour-derived slice cultures for the characterization of an EGFR-activating kinase mutation in a case of glioblastoma. BMC Cancer. 2018;18:964
72. Vilgelm AE, Saleh N, Shattuck-Brandt R, Riemenschneider K, Slesur L, Chen SC, Johnson CA, Yang J, Blevins A, Yan C. et al. MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci Transl Med. 2019;11:eaav7171
73. Zhang W, van Weerden WM, de Ridder CMA, Erkens-Schulze S, Schönfeld E, Meijer TG, Kanaar R, van Gent DC, Nonnekens J. Ex vivo treatment of prostate tumor tissue recapitulates in vivo therapy response. Prostate. 2019;79:390-402
74. Valta MP, Zhao H, Ingels A, Thong AE, Nolley R, Saar M, Peehl DM. Development of a realistic in vivo bone metastasis model of human renal cell carcinoma. Clin Exp Metastasis. 2014;31:573-584
75. Grosso SH, Katayama ML, Roela RA, Nonogaki S, Soares FA, Brentani H, Lima L, Folgueira MA, Waitzberg AF, Pasini FS. et al. Breast cancer tissue slices as a model for evaluation of response to rapamycin. Cell Tissue Res. 2013;352:671-684
76. Askoxylakis V, Ferraro GB, Kodack DP, Badeaux M, Shankaraiah RC, Seano G, Kloepper J, Vardam T, Martin JD, Naxerova K. et al. Preclinical Efficacy of Ado-trastuzumab Emtansine in the Brain Microenvironment. J Natl Cancer Inst. 2016;108:djv313
77. Minami N, Maeda Y, Shibao S, Arima Y, Ohka F, Kondo Y, Maruyama K, Kusuhara M, Sasayama T, Kohmura E. et al. Organotypic brain explant culture as a drug evaluation system for malignant brain tumors. Cancer Med. 2017;6:2635-2645
78. Zhang Y, Huang WJ, Yang QR, Zhang HD, Zhu XJ, Zeng M, Zhou X, Wang ZY, Li WJ, Jing HS. et al. Cryopreserved biopsy tissues of rectal cancer liver metastasis for assessment of anticancer drug response in vitro and in vivo. Oncol Rep. 2020;43:405-414
79. Nishida-Aoki N, Bondesson AJ, Gujral TS. Measuring Real-time Drug Response in Organotypic Tumor Tissue Slices. J Vis Exp. 2020
80. Lim CY, Chang JH, Lee WS, Lee KM, Yoon YC, Kim J, Park IY. Organotypic slice cultures of pancreatic ductal adenocarcinoma preserve the tumor microenvironment and provide a platform for drug response. Pancreatology. 2018;18:913-927
81. Jiang X, Seo YD, Sullivan KM, Pillarisetty VG. Establishment of Slice Cultures as a Tool to Study the Cancer Immune Microenvironment. Methods Mol Biol. 2019;1884:283-295
82. Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, Jalikis F, Yeung RS, Crispe IN, Pillarisetty VG. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology. 2017;6:e1333210
83. Schmid JO, Dong M, Haubeiss S, Friedel G, Bode S, Grabner A, Ott G, Mürdter TE, Oren M, Aulitzky WE, van der Kuip H. Cancer cells cue the p53 response of cancer-associated fibroblasts to cisplatin. Cancer Res. 2012;72:5824-5832
84. Rodriguez AD, Horowitz LF, Castro K, Kenerson H, Bhattacharjee N, Gandhe G, Raman A, Monnat RJ, Yeung R, Rostomily RC, Folch A. A microfluidic platform for functional testing of cancer drugs on intact tumor slices. Sci Transl Med. 2020;20:1658-1675
85. Voabil P, de Bruijn M, Roelofsen LM, Hendriks SH, Brokamp S, van den Braber M, Broeks A, Sanders J, Herzig P, Zippelius A. et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med. 2021;27:1250-1261
86. Ciraku L, Moeller RA, Esquea EM, Gocal WA, Hartsough EJ, Simone NL, Jackson JG, Reginato MJ. An Ex vivo Brain Slice Model to Study and Target Breast Cancer Brain Metastatic Tumor Growth. J Vis Exp. 2021
87. Philouze P, Gauthier A, Lauret A, Malesys C, Muggiolu G, Sauvaigo S, Galmiche A. CD44, γ-H2AX, and p-ATM Expressions in Short-Term Ex vivo Culture of Tumour Slices Predict the Treatment Response in Patients with Oral Squamous Cell Carcinoma. Int J Mol Sci. 2022;23:877
88. Das A, Alshareef M, Martinez Santos JL, Porto GBF, McDonald DG, Infinger LK, Vandergrift WA 3rd, Lindhorst SM, Varma AK, Patel SJ, D C. Evaluating anti-tumor activity of palbociclib plus radiation in anaplastic and radiation-induced meningiomas: pre-clinical investigations. Clin Transl Oncol. 2020;22:2017-2025
89. Alonso MC, Tabernero JM, Ojeda B, Llanos M, Solà C, Climent MA, Seguí MA, López JJ. A phase III randomized trial of cyclophosphamide, mitoxantrone, and 5-fluorouracil (CNF) versus cyclophosphamide, adriamycin, and 5-fluorouracil (CAF) in patients with metastatic breast cancer. Breast Cancer Res Treat. 1995;34:15-24
90. Leone JP, Leone J, Vallejo CT, Pérez JE, Romero AO, Machiavelli MR, Romero Acuña L, Domínguez ME, Langui M, Fasce HM. et al. Sixteen years follow-up results of a randomized phase II trial of neoadjuvant fluorouracil, doxorubicin, and cyclophosphamide (FAC) compared with cyclophosphamide, methotrexate, and 5-fluorouracil (CMF) in stage III breast cancer: GOCS experience. Breast Cancer Res Treat. 2014;143:313-323
91. Martín M, Ruiz A, Ruiz Borrego M, Barnadas A, González S, Calvo L, Margelí Vila M, Antón A, Rodríguez-Lescure A, Seguí-Palmer MA. et al. Fluorouracil, doxorubicin, and cyclophosphamide (FAC) versus FAC followed by weekly paclitaxel as adjuvant therapy for high-risk, node-negative breast cancer: results from the GEICAM/2003-02 study. J Clin Oncol. 2013;31:2593-2599
92. Bontenbal M, Creemers GJ, Braun HJ, de Boer AC, Janssen JT, Leys RB, Ruit JB, Goey SH, van der Velden PC, Kerkhofs LG. et al. Phase II to III study comparing doxorubicin and docetaxel with fluorouracil, doxorubicin, and cyclophosphamide as first-line chemotherapy in patients with metastatic breast cancer: results of a Dutch Community Setting Trial for the Clinical Trial Group of the Comprehensive Cancer Centre. J Clin Oncol. 2005;23:7081-7088
93. Hutchins LF, Green SJ, Ravdin PM, Lew D, Martino S, Abeloff M, Lyss AP, Allred C, Rivkin SE, Osborne CK. Randomized, controlled trial of cyclophosphamide, methotrexate, and fluorouracil versus cyclophosphamide, doxorubicin, and fluorouracil with and without tamoxifen for high-risk, node-negative breast cancer: treatment results of Intergroup Protocol INT-0102. J Clin Oncol. 2005;23:8313-8321
94. Lee HH, Wang YN, Xia W, Chen CH, Rau KM, Ye L, Wei Y, Chou CK, Wang SC, Yan M. et al. Removal of N-Linked Glycosylation Enhances PD-L1 Detection and Predicts Anti-PD-1/PD-L1 Therapeutic Efficacy. Cancer Cell. 2019;36:168-178.e164
95. He L, Lv Y, Song Y, Zhang B. The prognosis comparison of different molecular subtypes of breast tumors after radiotherapy and the intrinsic reasons for their distinct radiosensitivity. Cancer Manag Res. 2019;11:5765-5775
96. Xie G, Dong H, Liang Y, Ham JD, Rizwan R, Chen J. CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine. 2020;59:102975
97. Schubert ML, Schmitt M, Wang L, Ramos CA, Jordan K, Müller-Tidow C, Dreger P. Side-effect management of chimeric antigen receptor (CAR) T-cell therapy. Ann Oncol. 2021;32:34-48
98. Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD. et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med. 2018;378:439-448
99. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, Jäger U, Jaglowski S, Andreadis C, Westin JR. et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N Engl J Med. 2019;380:45-56
100. Kersting D, Fasbender S, Pilch R, Kurth J, Franken A, Ludescher M, Naskou J, Hallenberger A, Gall CV, Mohr CJ. et al. From in vitro to ex vivo: subcellular localization and uptake of graphene quantum dots into solid tumors. Nanotechnology. 2019;30:395101
101. Merz L, Höbel S, Kallendrusch S, Ewe A, Bechmann I, Franke H, Merz F, Aigner A. Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur J Pharm Biopharm. 2017;112:45-50
Author contact
![]() Corresponding author: Chu-Xia Deng, Ph.D. Dean and Chair Professor, E12, Room 4041, Faculty of Health Sciences, University of Macau, Macau SAR, China. Phone: (853) 8822 4997; Fax: 8822 2314; E-mail: cxdengedu.mo.
Corresponding author: Chu-Xia Deng, Ph.D. Dean and Chair Professor, E12, Room 4041, Faculty of Health Sciences, University of Macau, Macau SAR, China. Phone: (853) 8822 4997; Fax: 8822 2314; E-mail: cxdengedu.mo.

 Global reach, higher impact
Global reach, higher impact