Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(6):1664-1680. doi:10.7150/ijbs.78864 This issue Cite
Research Paper
Cardiovascular-related proteomic changes in ECFCs exposed to the serum of COVID-19 patients
1. Biomedicine, Biotechnology and Public Health Department, University of Cadiz, 11002 Cadiz, Spain
2. Biomedical Research and Innovation Institute of Cadiz (INiBICA), 11002 Cadiz, Spain
3. Department of Biochemistry and Molecular Biology, University of Southern Denmark, 5230 Odense, Denmark
4. Automation Engineering, Electronics and Computer Architecture and Networks Department, University of Cadiz, 11009 Cadiz, Spain
5. Department of Nursing, National Paraplegic Hospital, Toledo, Spain
6. Occupational Health Service, National Paraplegic Hospital, SESCAM, 45071, Toledo, Spain
7. Internal Medicine Department, University Hospital Virgen del Rocío, Seville, Spain
8. UGC Microbiología, University Hospital Puerta del Mar, Avda. Ana de Viya 21, 11009, Cádiz, Spain
9. Cell Biology, Physiology and Immunology Department, Agrifood Campus of International Excellence (ceiA3), University of Cordoba, 14014, Córdoba, Spain
10. Maimonides Biomedical Research Institute of Cordoba (IMIBIC), UGC Nephrology, Hospital Universitario Reina Sofia, 14004, Cordoba, Spain
11. Psychology Department, University of Cádiz, 11510, Puerto Real, Spain
12. Biomedical Research Networking Center for Mental Health Network (CIBERSAM), Institute of Health Carlos III, 28029, Madrid, Spain
13. Laboratory of Neuroinflammation, National Paraplegic Hospital, 45071 Toledo, Spain
14. Redes de investigación cooperativa orientadas a resultados en salud, Enfermedades vasculares cerebrales, RICORS-ICTUS, SESCAM
*Both authors have equally contributed to this work.
Received 2022-9-12; Accepted 2023-2-2; Published 2023-3-5
Abstract
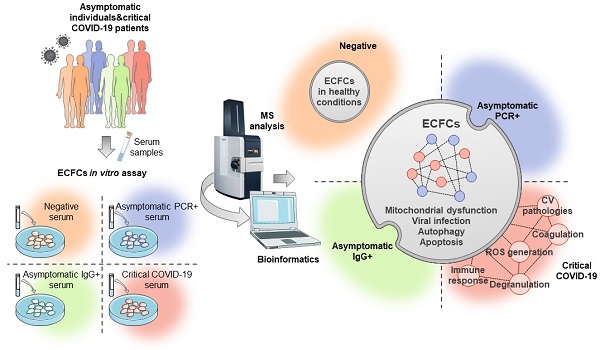
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection significantly affects the cardiovascular system, causing vascular damage and thromboembolic events in critical patients. Endothelial dysfunction represents one of the first steps in response to COVID-19 that might lead to cardiovascular complications and long-term sequelae. However, despite the enormous efforts in the last two years, the molecular mechanisms involved in such processes remain poorly understood. Herein, we analyzed the protein changes taking place in endothelial colony forming cells (ECFCs) after the incubation with the serum from individuals infected with COVID-19, whether asymptomatic or critical patients, by application of a label free-quantitative proteomics approach. Specifically, ECFCs from healthy individuals were incubated ex-vivo with the serum of either COVID-19 negative donors (PCR-/IgG-, n:8), COVID-19 asymptomatic donors at different infective stages (PCR+/ IgG-, n:8and PCR-/IgG+, n:8), or hospitalized critical COVID-19 patients (n:8), followed by proteomics analysis. In total, 590 proteins were differentially expressed in ECFCs in response to all infected serums. Predictive analysis highlighted several proteins like CAPN5, SURF4, LAMP2 or MT-ND1, as highly discriminating features between the groups compared. Protein changes correlated with viral infection, RNA metabolism or autophagy, among others. Remarkably, the angiogenic potential of ECFCs in response to the infected serums was impaired, and many of the protein alterations in response to the serum of critical patients were associated with cardiovascular-related pathologies.
Keywords: COVID-19, ECFCs, Viral infection, RNA metabolism, cardiovascular diseases, endothelial dysfunction, autophagy, proteomics, mass spectrometry
Background
Coronavirus disease 2019 (COVID-19) was declared as a global pandemic on March 11, 2020, because of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, provoking more than 6,7 million deaths worldwide (www.covid19.who.int). SARS-CoV-2 not only affects the respiratory system, it also provokes severe vascular damage as well as thromboembolic events responsible for many associated clinical complications [1, 2]. Importantly, in relation to the former processes, endothelial dysfunction plays a significant role in the pathogenesis of COVID-19, either by direct infection through SARS-CoV-2 [3] or as result of the activation of inflammatory leukocytes promoted by the vascular endothelium, leading to a cytokine storm responsible for systemic inflammation [4-6].
Although the effects are more potentiated in severe patients, endothelial dysfunction takes place even in asymptomatic individuals [7], and different signs of persistent endothelial activation and related inflammation can be found even in convalescent COVID-19 patients, including elevated levels of circulating endothelial cells (CECs) [8, 9]. Hence, a better understanding of the initial stages in which SARS-CoV-2 affects the endothelium is required, in order to predict or prevent unwanted secondary effects, and the risk of suffering from long-term cardiovascular (CV) complications [5, 7].
To date, different approaches have arisen to evaluate the potential mechanisms by which SARS-CoV-2 might promote endothelial damage, from in vitro studies with human organoids and organ-on-chip platforms [4, 10-12], to in vivo assays with animals expressing ACE2 and further infected with SARS-CoV-2 [13]. Very recently, we described a cell model to analyze the response of vascular cells to SARS-CoV-2 infection, based on the incubation of circulating angiogenic cells (CACs), also considered as CECs, with the serum of COVID-19 asymptomatic donors, identifying many proteins related to endothelial dysfunction and inflammatory response after viral infection, corroborating the potential of this approach [7]. In the current study, we have extended our strategy by evaluating the effect of the serum factors from asymptomatic to critical COVID-19 patients over endothelial colony forming cells (ECFCs) isolated from white adipose tissue of healthy adults, by application of advanced mass spectrometry (MS)-based proteomics methods. We and other researchers have already shown that these cells present a robust clonogenic and proliferative potential and they are known to promote vascular repair [14-18]. On the other hand, under pathological environments, ECFCs become dysfunctional; moreover, in response to adverse conditions, impaired ECFCs might contribute to the endothelial dysfunction related to cardiovascular diseases (CVDs) [19-22]. Remarkably, elevated levels of ECFCs have been found in 3 months post-COVID patients [9]. Thus, ECFCs may not only provide novel opportunities in identifying biomarker of post-COVID endothelial damage, but also represent an optimal candidate to tackle SARS-CoV-2 endothelial infection and a platform to evaluate therapeutic strategies against the disease.
Methods
Serum sample acquisition
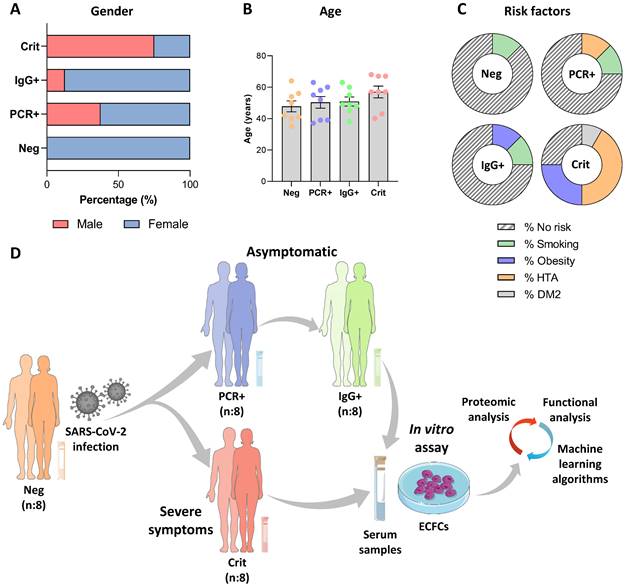
The study was conducted with COVID-19 negative donors and asymptomatic individuals (PCR+/IgG- and PCR-/IgG+) recruited at the National Paraplegic Hospital (SESCAM), Toledo, Spain during April-May 2020 and critical COVID-19 patients admitted at the COVID-19 hospital, Seville, Spain, and the Puerta del Mar University Hospital, Cádiz, Spain, during May-June 2021. Donor characteristics are shown in Figure 1A-C. A SARS-CoV-2 qPCR analysis from nasopharyngeal samples and an ELISA assay testing for specific IgG and IgM antibodies (IME00136 and IME00137; Erba Mannheim) were performed to determine their status. Briefly, peripheral blood samples were collected using serum separator tubes (SSTTM II advance, BD Vacutainer®), centrifuged and stored at -80 ºC, as described [7].
Donors were classified in four different groups: COVID-19 negative donors (Neg, n:8), with both qPCR and antibody's test (PCR-/IgG-), asymptomatic patients PCR positive (PCR+, n:8) or IgG positive (IgG+, n:8) at the time of blood collection, and critical COVID-19 patients that required hospitalization (Crit, n:8) (Figure 1D).
ECFCs isolation and culture
ECFCs were isolated from normal subcutaneous white adipose tissue and cultured as previously described [18]. ECFCs were purified by magnetic activated cell sorting using CD31-coated magnetic beads, plated in 1% gelatin coated plates and incubated in EBM-2 media plus 20% FBS and Single Quots growth factors (except for hydrocortisone) (Lonza). ECFCs were characterized in vitro and in vivo as described [18].
ECFCs incubation ex vivo with patients' serum
ECFCs were washed several times with PBS 1X, to discard any remaining traces of FBS from the initial conditioned media, and then incubated for 24 h (37 ºC, 5% CO2) with EBM-2 medium containing 10% serum of the Neg, PCR+, IgG+ and Crit groups (n:8 per group), as described [7]. Cells were collected using Trypsin-EDTA 1X (X0930-100; Biowest), centrifuged and washed once with PBS 1X.
Study population characteristics and schematic representation of the experimental assay. Graphical representation of the donors' A) gender, B) age and C) risks factors for each group. D) Schematic representation of the experimental assay. The serums from COVID-19 patients in different stages, including negative, asymptomatic and critical individuals were collected: SARS-CoV-2 negative (PCR-/IgG-, n:8) and SARS-CoV-2 positive, at the peak of infection (PCR+/IgG-), asymptomatic (n:8) and critical (n:8), or after the infective peak (PCR -/IgG +, n:8). Next, ECFCs from healthy donors were incubated with all four sets of serum samples and a label free quantitative approach was performed, followed by bioinformatics analysis (statistic and functional classification).
Proteomic analysis
The proteome changes of ECFCs in response to the incubation with the different sets of serum samples (Neg n:8; PCR+ n:8; IgG+ n:8; Crit n:8) were analyzed using tandem mass spectrometry (LC-MS/MS)-based label free quantitative (LFQ) analysis. Briefly, proteins were extracted by resuspending ECFCs pellets in 8M urea containing protease inhibitors (04693132001; Roche) and their amount was estimated using Qubit Fluorometric system (ThermoFisher Scientific) following manufacturer´s guidelines. Next, 50 µg of proteins in 8M urea per sample were reduced (10 mM Dithiothreitol) and alkylated (50 mM Iodoacetamide), and then diluted four times with 50 mM ammonium bicarbonate prior to digestion with Trypsin/LysC (V5073; Promega) (enzyme/substrate ratio 1:50), at 37 °C, overnight. Finally, digestion was quenched with 0.1% TFA before peptide purification with C18 micro-columns, as described [23], and eluates were dried with a speed-vac system.
Digested peptides were reconstituted in 0.1% formic acid (FA) and a NanoDrop (DeNovix, DS-11 Spectrophotometer) was used to estimate the peptide concentration. An amount equivalent to 200 ng of peptides were analyzed on a timsTOF Pro mass spectrometer (Bruker Daltonics) interfaced with Easy nLC (Thermo Scientific). Briefly, peptides were separated on an Aurora Series UHPLC emitter column (250 mm X 75 µm id, 1.6 µm C18) from IonOpticks, using the solvents A (0.1% FA) and B (95% acetonitrile with 0.1% FA). Peptides were eluted at flow rate of 300 nL/min, with increasing % of solvent B from 5 to 45% in 60 min (from 5 to 25% in 50 min followed by increasing to 45% in 10 min) and further column washing with 95% B. The column temperature was maintained to 45 ºC. The instrument was operated in diaPASEF mode via Captive nano-electrospray source (Bruker Daltonics) at 1400 V with an accumulation time of 100 ms and a ramp of 100 ms. A 25 m/z precursor isolation width was used to cover 400 to 1200 m/z, covering an ion mobility range /1/K0) from 0.60 to 1.60 V.s/cm2 [24].
Data processing and statistics
diaPASEF files were analyzed using Spectronaut (v 15.2.210819.50606, Biognosys AG) in directDIA™ mode with default settings except protease selected were Trypsin and LysC. The detailed description of all the parameters can be found in Table S8.
The Spectronaut output was then exported into tabular format for further analysis. In the Perseus software [25], protein intensity values were log2 transformed and samples were categorically annotated to define the conditions. A t-test differential expression analysis was used with a permutation-based FDR calculation. Proteins were considered as differentially expressed between the groups (Neg, PCR+, IgG+ and Crit) when FDR < 0.05 and log2 foldchange > 1 (up-regulated) or < -1 (down-regulated). These changes were confirmed afterwards with GraphPad Prism 9 software. Data were presented as box plots graphs representing median, min and max value and showing all points.
Additional data processing was done using Venny v2.1[26], Python, R and MetaboAnalyst [27]. Specifically, Clustergrammer was used to generate an interactive heatmap [28]. The functional role of proteins was analyzed using Ingenuity Pathway Analysis (IPA) software, Reactome (https://reactome.org/), Erichr (https://maayanlab.cloud/Enrichr/), String (https://string-db.org) and Coronascape (COVID-19 Reference Gene Lists (metascape.org).
Machine learning algorithms
We explored the potential of machine learning techniques to classify ECFCs treated with the serum from asymptomatic (PCR+, IgG+) or COVID-19 negative individuals, and from critical COVID-19 patients. Due to the limited generalizability of the results imposed by the small size of the data set, three low complexity models were used: MLR, NB and LSVM. Feature selection was used to identify proteins with high discriminant power to reduce the high dimensionality of the data set [29] and circumvent the so-called curse of dimensionality. Attribute sets were evaluated by using a wrapper 5-fold cross validation learning scheme. Area under the ROC curve (AUC) was used to assess the performance of attribute combinations. The space of attribute subsets was bidirectionally searched by greedy hill climbing augmented with a backtracking facility. The number of consecutive non-improving nodes to allow before terminating the search was fixed at 5. The following metrics of performance for each classifier were calculated using 5-fold cross validation: accuracy, AUC, true and false positive rates, recall and Kappa statistic. WEKA data mining software [30] and MATLAB (The MathWorks Inc., Natick, USA) were used for building the models.
In silico interaction analysis between serum and ECFCs altered proteins
In order to predict serum factors potentially responsible of the changes seen in ECFCs, we conducted an exhaustive literature search, selecting proteomic studies identifying proteins altered in the serum of asymptomatics or critical COVID-19 patients (Table S9). Next, an in-silico analysis was performed with Fluorish software (Flourish | Data Visualization & Storytelling), evaluating potential interactions between the proteins altered in the serum of COVID-19 patients' vs negative controls (detected at least in 2 of the selected articles) with the protein changes identified in ECFCs in response to the serum from COVID-19 patients.
Angiogenesis assay
A tube formation assay was performed to evaluate the effect of the serum from COVID-19 patients over the ECFCs angiogenic potential. ECFCs (15000 cells/well) were seeded into a 96 well angiogenesis μ-plate (Ibidi, 81506) pre-coated with 10 μl matrigel (Bioscience, 356231), as described [31], and incubated for 24 h (37 ºC, 5% CO2) with EBM-2 medium containing 10% serum of the Neg, PCR+, IgG+ and Crit groups (n:4 per group), in triplicates. In addition, ECFCs were incubated with 35 ng/ml Fibroblast Growth Factor (FGF, R&D Systems) and 15 mM sulforaphane (S4441-5MG, Sigma) as angiogenic activator and inhibitor controls respectively. After 24 hours, images were taken per well with an inverted phase-contrast microscope and the number of meshes were quantified.
Anti-inflammatory assay
ECFCs were seeded in 6-well plates, previously coated with 1% gelatin, at a density of 30000 cells/cm2 in EBM-2 medium and 10% FBS. After 12 h of incubation (37 ºC, 5% CO2), the medium was replaced, and cells were incubated with EBM-2 medium containing 10% of serum from the Neg, PCR+, IgG+ and Crit groups (n:3 per group). In addition, another set of cells were incubated with EBM-2 with 2% FBS with or without 10 ng/ml of tumor necrosis factor-α (TNF-α; R&D Systems), as negative and positive controls of the inflammatory response respectively. All wells were incubated (37 ºC, 5% CO2) for another 5 hours. Cells were then detached and washed with cytometry buffer (1x PBS, 2.5% FBS, 2 mM EDTA), and then incubated with anti-human VCAM-1 antibody (305809; Biolegend) for 20 min at 4ºC in the dark. Samples were analyzed using CytoFLEX cytometer (Beckam Coulter, USA) and CytExpert software. Finally, data was analyzed with FlowJo v10.4 software.
Results
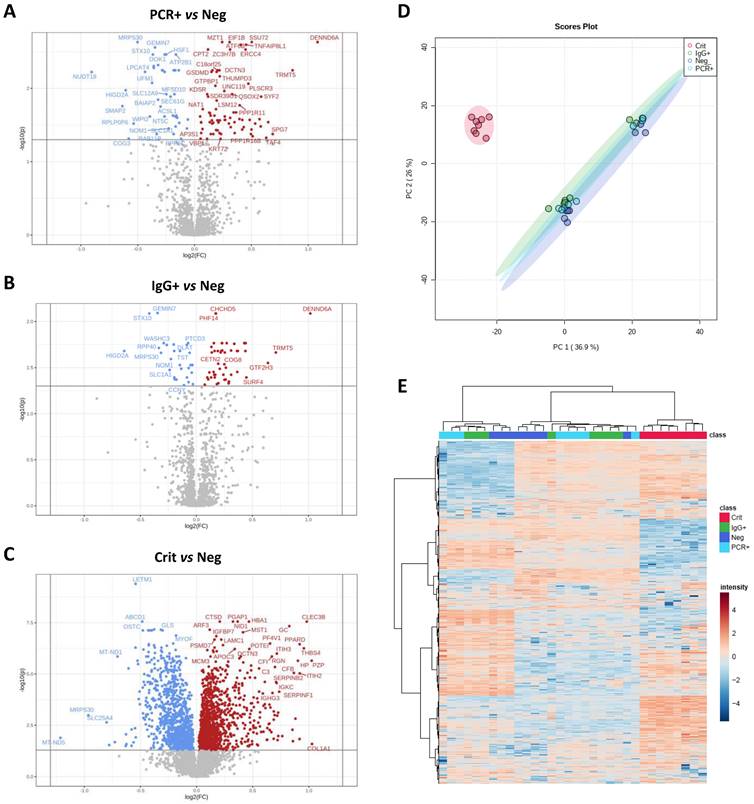
A total of 5052 proteins (4883±129 on average) were identified in ECFCs treated with the serum from COVID-19 negative (ECFCs+Neg, n:8), PCR+ (ECFCs+PCR, n:8) and IgG+ (ECFCs+IgG, n:8) asymptomatics, and critical COVID-19 patients (ECFCs+Crit, n:8). The statistical analysis identified 590 differentially expressed proteins between these groups (Figure 2A-C). The proteome profile of ECFCs incubated with the serum of critical COVID-19 patients was clearly different than the rest of conditions, as indicated by the principal component analysis (PCA, Figure 2D) and the hierarchical clustering classification (Figure 2E). Furthermore, compared to the other groups, ECFCs+Crit reported the highest number of protein changes (up and down-regulated) (Figure 3A and B). Nevertheless, protein differences were also found in ECFCs treated with the serum of asymptomatic (PCR+ and IgG+) vs ECFCs+Neg control donors. Full information regarding identification and quantification data (log2 fold changes and p-values) can be found in Tables S1-4.
ECFCs proteomic changes in response to the serum of COVID-19 patients in different stages and symptomatology. Volcano plots representative of protein up (red) and down-regulated (blue) in ECFCs incubated with the serum of A) PCR+ vs Neg donors, B) IgG vs Neg and C) Crit vs Neg. D) Principal component analysis. E) Hierarchical cluster representing the differential protein profiles. Interactive heatmap available as a supplementary file in html format.
Common protein changes in ECFCs in response to asymptomatic and critical COVID-19 serum. A) Representation of the number of proteins up- (red) and down-regulated (blue) in ECFCs incubated with the serum of asymptomatic (PCR+, IgG+) or Crit patients compared to ECFCs + Neg. B) Venn's diagram showing the overlapping of proteins differently expressed between ECFCs incubated with the serum of asymptomatic (PCR+, IgG+) or Crit compared to ECFCs + Neg. From the 25 common proteins altered in ECFCs incubated with the serum of infected donors (PCR+, IgG+ and Crit) compared with ECFCs+Neg, the proteins up- C) or down-regulated D) are shown, including representative graphs with the LFQ intensities registered for some of the proteins highlighted by predictive analysis. *p-value<0.05, **p-value<0.01, ***p-value<0.001, ****p-value<0.0001.
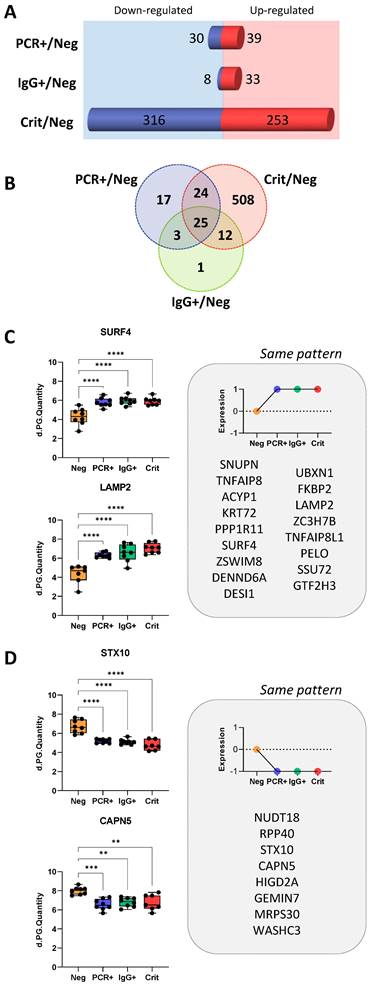
Common protein changes in ECFCs in response to asymptomatic and critical COVID-19 serum
From the total of 590 differentially expressed proteins, 508 were identified only in ECFCs+Crit vs ECFCs+Neg cells, while 24 of them were commonly altered between ECFCs+Crit and ECFCs+PCR, and 25 proteins were commonly altered in response to the serum factors of infected individuals, COVID-19 asymptomatic (PCR+ and IgG+) donors and critical patients, compared with ECFCs+Neg (Figure 3A-D).
According to Reactome Pathway Database (Table S5), the 25 common proteins up- or downregulated in ECFCs exposed to the serum of infected individuals, were mostly related to the metabolism of RNA (RPP40, GTF2H3, SNUPN, GEMIN7) and also non-coding RNA (SNUPN, GEMIN7), including RNA polymerase activity and regulation (GTF2H3), intra-Golgi and Golgi-to-ER trafficking (SURF4, STX10), as well as chaperone mediated autophagy (LAMP2). In addition, the analysis revealed that some of these proteins had been previously associated to SARS-CoV-2 early (FKBP2; UBXN1; PPP1R11), middle (UBXN1; PPP1R11; CAPN5) and late-stage infection in human male blood (FKBP2; UBXN1; PPP1R11; SURF4; SSU72).
Therefore, these protein changes seem relevant since they remained even when ECFCs were exposed to the serum of individuals that had overcome the infection with no apparent symptoms (ECFCs+IgG).
Differential protein expression patterns in ECFCs exposed to asymptomatic serums
Focusing on the response of ECFCs to asymptomatic serums, our results indicated that the expression patterns of ECFCs+PCR or ECFCs+IgG cells were similar, compared with ECFCs exposed to the serum of COVID-19 negative donors (Figure 4A). Some of these changes are shown in Figure 4B.
According to KEGG pathways, proteins altered in ECFCs+PCR or ECFCs+IgG appeared to participate, among others, in mRNA surveillance (SSU72, PELO) and RNA transport (RPP40, GEMIN7, SNUPN), and also related to phagosomes (ITGB5, SEC61G, LAMP2) or autophagy (CTSL, LAMP2, WIPI2) (Figure 4C). Similarly, the Ingenuity Pathway Analysis (IPA) classification platform connected several proteins up- and downregulated in response to the asymptomatic serum factors to viral infection, apoptosis and autophagy (Table S6) and, in the case of the ECFCs+PCR group, protein changes were specifically associated with severe acute respiratory syndrome (ACSL1, MZT1, ZSWIM8) (Figure 4D).
Proteomic changes in ECFCs exposed to asymptomatic serums. A) Hierarchical cluster representing the differential protein profiles for ECFCs incubated with the serum of asymptomatic (PCR+, IgG+) and COVID-19 Neg donors. B) Graphical representation of the LFQ intensities registered for proteins altered in ECFCs exposed to asymptomatic serums. C) Altered pathways in ECFCs stimulated with serum from asymptomatic donors compared to ECFCs+Neg cells. The p-values (-log10) obtained by Kegg Pathway platform and the calculated SEM are represented. D) IPA functional network with proteins up- (red) or down-regulated (green) in PCR+ vs Neg correlated with viral infection and severe acute respiratory syndrome. *p-value<0.05, **p-value<0.01, ***p-value<0.001, ****p-value<0.0001.
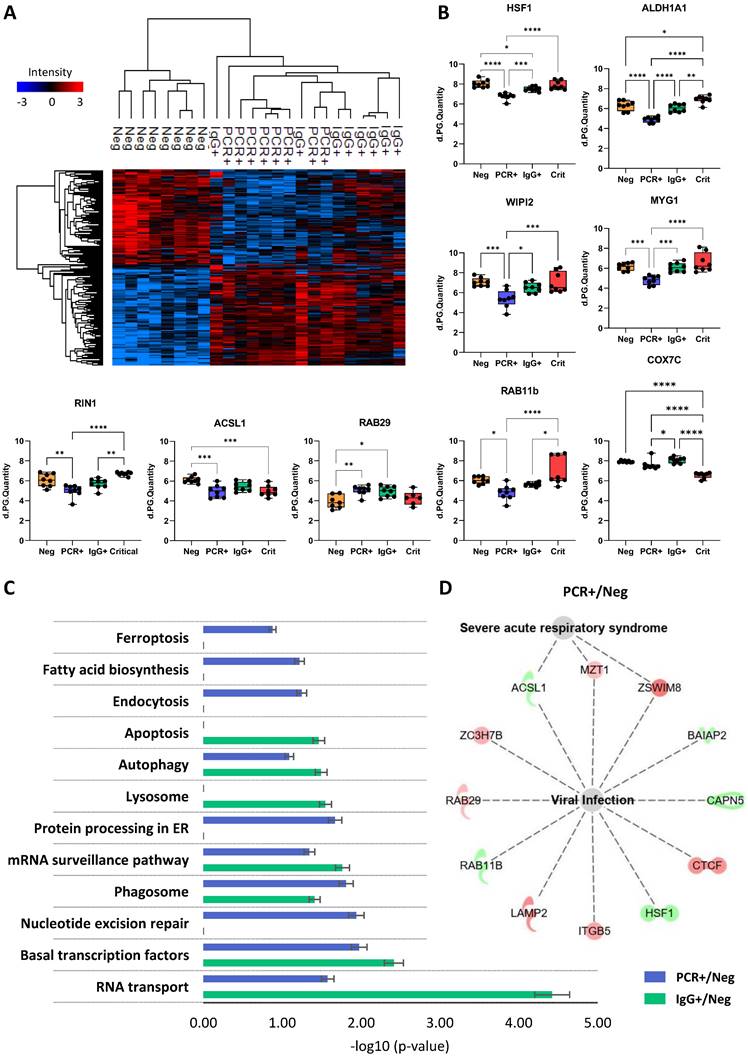
These data corroborate our initial findings suggesting that SARS-CoV-2 promotes molecular alterations even in total or partial absence of classical symptoms [7], and that these changes persist when the infection has disappeared (PCR-/IgG+).
Proteomic changes in ECFCs in response to the serum of critical patients
As indicated above, most changes were seen in ECFCs in response to the serum of COVID-19 critical patients. For example, proteins like S100A11, PF4, PPARD, MIF, THBS4, or ITGB5, were up-regulated in ECFCs+Crit compared to the other groups (Figure 5A). Based on the results provided by “coronascape” platform [32], supported itself by several database online platforms such as Reactome, Go Biological or KEGG pathways, the proteins differentially expressed in ECFCs+Crit were involved in many different pathways associated to viral infection, including RNA metabolism, ribonucleoprotein complex biogenesis, signaling by RhoGTPAses, or vesicle mediated transport, as well as SARS-CoV and human immunodeficiency virus (HIV) infections, neutrophil degranulation or cellular response to stress, among others (Figure 5B and Figure S1). More directly, IPA platform correlated the protein changes detected in ECFCs (Crit vs Neg) with RNA virus and COVID-19 infection (Figure 5C), as well as with functions altered or up-regulated in severe COVID-19 patients, such as immune response, coagulation, infection and apoptosis (Table 1).
Functional classification of differentially expressed proteins in ECFCs incubated with critical COVID-19 patient's serum (Crit) vs incubated with healthy donor's serum (Neg). Protein classification was made with the IPA software based on biomedical literature and integrated databases. The table shows the most probable functions in which the proteins of interest are involved, p-value, activation z-score, gene names and number
| Functions | P-value | Z-score | Gene names | Number of IDs |
|---|---|---|---|---|
| Immune response of leukocytes | 9.95E-04 | 2.441 | APCS, APOA1, APOA2, APOE, BCL2L1, C3, CD59, HSDL1, IGHG3, IGHM, IL1RL1, IRF3, ISG15, LGALS9, LTF, MAPKAPK2, MIF, MRTFA, PF4, PLAUR, PLCG2, SOAT1, TCIRG1 | 23 |
| Degranulation | 1.23E-08 | 1.672 | A2M, ABCC4, ACAA1, ADAM10, AHSG, ALB, APOA1, APOH, APOOL, C3, CD59, CLEC3B, CMTM6, CTSD, CYB5R1, DHCR7, F2, F5, FERMT3, FGG, FTH1, HBB, HP, ITGAE, ITIH3, KRT1, LAMP2, LGALS9, LTF, MAGT1, METTL7A, MIF, NCSTN, NDUFC2, ORMDL3, PF4, PLAUR, PLCG2, PLG, PPP3CC, PSMB7, RICTOR, S100A11, STING1, SURF4, TCIRG1, TMEM179B, TTN | 48 |
| Coagulation | 3.46E-04 | 0.751 | A2M, APOE, APOH, BLOC1S6, C3, CD59, COL1A1, EHD3, F2, F5, FGG, GNA11, GNAQ, HBB, HP, MRTFA, PF4, PLAUR, PLCG2, PLG, SERPINF1 | 21 |
| Viral Infection | 1.13E-05 | 0.376 | ACSL1, ADAM10, AFG3L2, AHSG, ALB, AP1S1, AP1S2, APCS, APOA1, APOA2, APOB, APOBEC3F, APOE, APOH, BAIAP2, BCL2L1, C3, C4BPA, CAPN5, CCDC51, CFB, CFI, CLEC3B, COL5A1, CTCF, CTSL, DYSF, ELOVL5, F2, F5, FMR1, GCAT, GNAQ, GORASP1, GPAT3, GRIN3A, HBB, HP, IGHM, IRF3, ISG15, ITGB5, ITIH3, KIF3A, KRT8, LAMP2, LGALS9, LONP1, LRPPRC, LTF, MAGT1, MARCHF2, MCL1, MIF, MTX1, MX1, MZT1, NLRX1, PARP12, PCLO, PCYOX1, PF4, PLCG2, PLG, PPAN, PPP3CC, PSMB9, PTDSS1, RICTOR, RPL3, RPL38, RPL5, RPS6, RTN3, RXRB, SFXN3, SOAT1, STING1, TCIRG1, TLE5, TOP2A, TOP2B, TRIM22, XAF1, ZC3H7B, ZFP36L2, ZSWIM8 | 87 |
| Cell Infection | 8.30E-04 | 0.225 | AFG3L2, APCS, APOE, APOH, C3, CAPN5, CCDC51, CFI, COL5A1, CTSL, F2, GNAQ, GORASP1, IRF3, KIF3A, LGALS9, LRPPRC, LTF, MX1, NLRX1, PTDSS1, RPL3, RPL5, RPS6, RTN3, SFXN3, TLE5, ZC3H7B | 28 |
| ROS Generation | 2.14E-05 | -0.612 | AATF, ALB, ALDH2, APOA1, APOE, BCL2L1, BNIP3, DHCR24, F2, HP, ITM2B, LTF, MACROH2A1, MIF, PLAUR, PPARD, SERPINF1, SOD2, TXNRD1 | 19 |
| Necrosis/Apoptosis | 1.98E-05 | -1.181 | A2M, AATF, ABCC4, ABCG2, ACP1, ACSL1, ADAM10, ALB, ALDH2, APOA1, APOB, APOC3, APOE, ARMC10, BCL2L1, BNIP3, C3, CD59, CDCA2, CFB, CHP1, COL1A1, CS, CTCF, CTSD, DAP3, DCK, DHCR24, DHCR7, DYSF, EBNA1BP2, ELOVL5, F2, F5, FAIM, FAP, FECH, FLT4, FMR1, FTH1, GLS, GLUD1, GNAQ, GNL3, GPNMB, GRIN3A, HADHA, HBB, HTRA2, IARS2, IDH2, IGHM, IL1RL1, IMMT, IRF3, ISG15, ITM2B, KIF3A, KRT8, LAMA4, LAMA5, LAMP2, LGALS9, LONP1, LRRC8A, LTF, MAPK7, MAPKAPK2, MCL1, MIF, MRTFA, MST1, MTCH2, MX1, MYOC, NCSTN, NDUFA13, NLRX1, ORMDL3, PAK4, PCK2, PEX11B, PF4, PLAUR, PLCG2, PLG, PMVK, POLR2H, PPARD, PPP1R11, PTPRE, PTPRF, PTRH2, RBM3, RFK, RGN, RICTOR, RNASEH2A, RPL27A, RPL3, RPL38, RPL5, RPL6, RPS24, RPS6, RRP1B, S100A11, SEC61G, SERPINB2, SERPINF1, SLC16A1, SLC1A1, SLC25A11, SLC25A4, SLC9A1, SOAT1, SOD2, SRPX, STING1, SURF1, TAF4, THBS4, TNFAIP8, TNFAIP8L1, TOP1, TOP2A, TOP2B, TRAP1, TTN, TXNRD1, TYMP, VTI1A, XAF1, ZMPSTE24 | 134 |
| Autophagy | 8.32E-04 | -1.306 | ABHD5, ACSL1, BAIAP2, BCL2L1, BNIP3, CTSD, CTSL, ERCC4, FOXK1, IRF3, LAMP2, LAMTOR4, LETM1, MCL1, NAF1, NLRX1, ORMDL3, PLG, RAB8A, RICTOR, SLC25A4, SLC9A1, SOAT1, SOD2, SRPX, STING1, TCIRG1, TOMM22, TOMM6, TRIM22, VTI1A, ZMPSTE24 | 32 |
| Mitochondrial Dysfunction | 1,81E+01 | ACO2, ATP5ME, ATP5MF, ATP5MG, ATP5PB, ATP5PD, ATPAF2, COX4I1, COX5B, COX6C, COX7B, COX7C, CPT1A, HTRA2, MT-CO3, MT-ND1, MT-ND5, NCSTN, NDUFA13, NDUFA9, NDUFB11, NDUFB3, NDUFB5, NDUFB6, NDUFB7, NDUFB8, NDUFS2, RHOT2, SOD2, SURF1, UQCR10 | 31 |
ECFCs differential protein expression in response to the serums of critical patients. A) Schematic representation of proteins altered (up or down-regulated) in ECFCs only after incubation with the with serums from critical patients. Also, representative graphs with the LFQ intensities registered for some of the altered proteins are shown. B) Hierarchical functional clustering provided by by Coronascape for proteins differentially expressed in ECFCs in response to critical COVID-19 serums (vs Neg). See Figure S1 for full information. C) IPA functional network including proteins up- (red) or down-regulated (green) in ECFCs+Crit vs ECFCs+Neg correlated with RNA virus and COVID-19 infection.
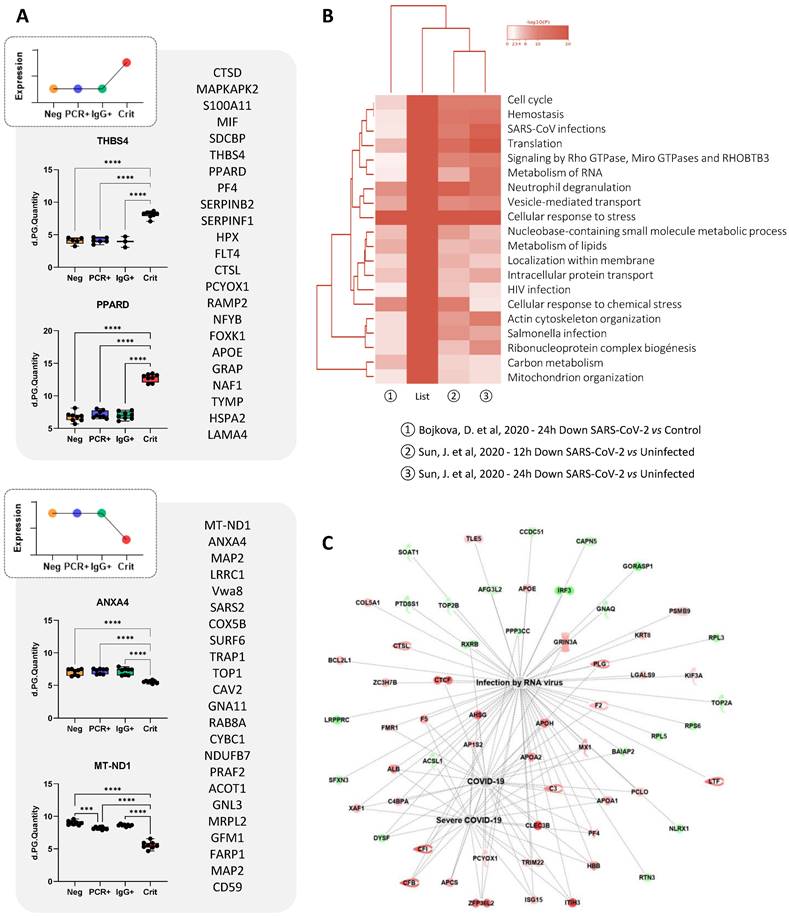
Discriminating proteins highlighted by machine learning prediction tools
A multinomial logistic regression (MLR), Näive Bayes (NB) and linear support vector machines (LSVM) classifiers were trained and cross-validated to automatically classify ECFCs+Neg, ECFCs+PCR, ECFCs+IgG and ECFCs+Crit samples. The application of these three machine learning techniques reported several proteins highly discriminating between the four groups. The performance of all classifiers was very promising, as evidenced by the estimated metrics. ALDH1A and MT-ND1 molecules were selected as relevant for the three predictive models while SDCBP, HTRA2, CAPN5, STX10, and RIN1 proteins appeared as features with high predictive value in two of the validated models (Table 2 and Figure S2).
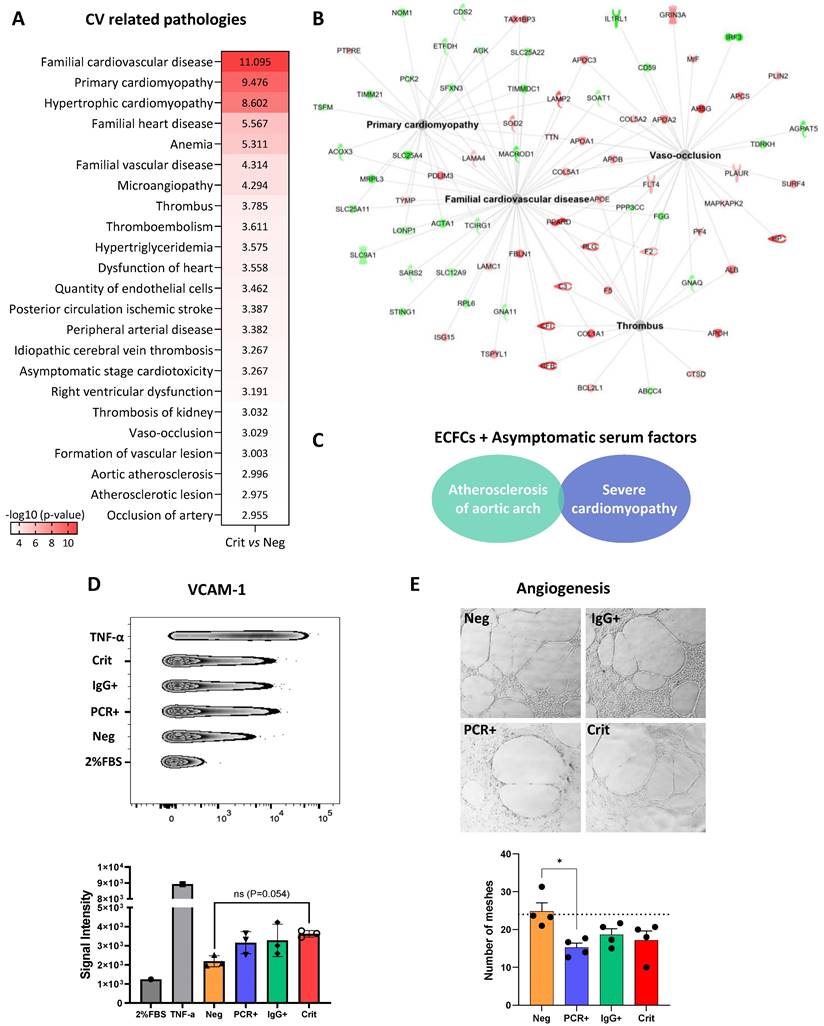
Cardiovascular related proteomic changes in ECFCs treated with the serum of infected individuals
Many of the protein changes seen in ECFCs incubated with the serum from critical patients (ECFCs+Crit) were associated with cardiovascular-related pathologies, including vaso-occlusion, atherosclerosis and thrombotic related processes, cardiomyopathy, ischemic stroke or peripheral artery disease among others (Figure 6A-B and Table S7). Among these, proteins like lysosome-associated membrane protein 2 (LAMP2) and surfeit locus protein 4 (SURF4) were over-expressed in ECFCs+Crit cells but also in ECFCs treated with sera from asymptomatics (ECFCs+PCR and ECFCs+IgG). Indeed, although to a much lesser extent, asymptomatic sera also promoted changes associated with CVDs such as atherosclerosis of aortic arch and severe cardiomyopathy (Figure 6C). In agreement with the proteomic results, functional assays reported an up-regulation of the vascular cell adhesion molecule (VCAM1), a marker of endothelial dysfunction and inflammation correlated with CVD [33], in response to all positive COVID-19 serums, whether from asymptomatics or critical patients (Figure 6D). Moreover, the angiogenic potential of ECFCs was impaired (Figure 6E).
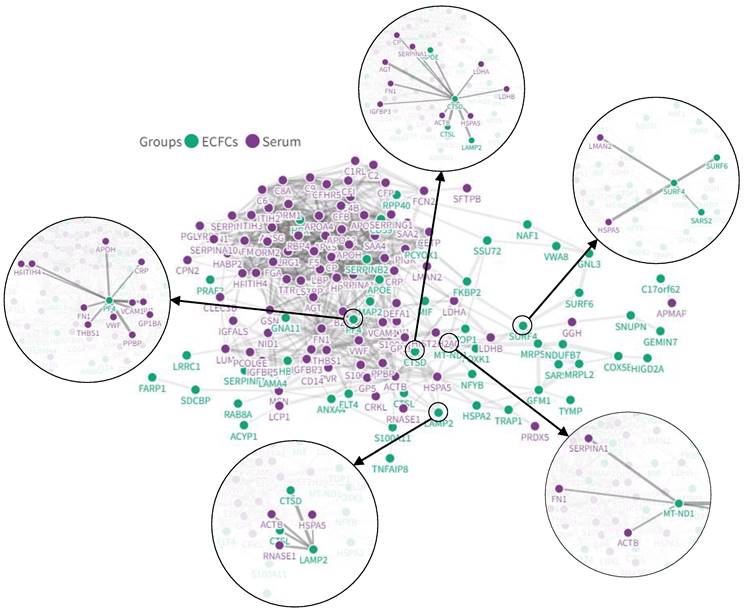
Interaction networks between COVID-19 serum and ECFCs altered proteins
Different serum proteins that have been reported as altered after COVID-19 infection (i.e. CRP, von Willebrand factor, HSPA5, or LMAN2) in several proteomic studies (Table S9) were directly connected with the protein changes seen in ECFCs in the current approach in response to COVID-19 positive sera (SURF4, LAMP2, PF4, CTSL or CTSD, among others) (Figure 7).
Discussion
At present, different serum markers associated to the severity of SARS-CoV-2 have been disclosed, including the C-reactive protein (CRP), procalcitonin (PCT), or ferritin [34], as well as the so-called “cytokine storm” (IL-6, TNF-α and other pro-inflammatory factors) [35], or markers directly related to coagulation (D-dimer) or cardiac injury, such as troponin, N-terminal (NT) proB-type natriuretic peptide (BNP), or creatine kinase (CK) [36, 37]. Additionally, serum miRNA targeting ACE2 or other genes have also been reported [38, 39]. Many of these biomarkers are indicators of tissue injury or even the severity of the disease, but they cannot explain the differential predisposition of individuals to respond against the infection, or why many individuals suffer from long-term sequelae, with no clear explanation yet of how COVID-19 symptoms remain. Therefore, it is still a long way to understand the mechanisms of action of SARS-CoV-2 as well as the organism´s response to this virus.
Endothelial dysfunction represents one of the most characteristic effects of SARS-CoV-2 and a major underlying mechanism responsible of CV complications in COVID-19 patients [40]. Indeed, critical patients present elevated levels of endothelial biomarkers compared to non-critical ones, suggesting a prognostic role of endothelial dysfunction in COVID-19 disease [41]. Furthermore, the presence of increased levels of ECFCs in 3 months post-COVID-19 patients compared to healthy subjects, might also represent a marker of post-COVID endothelial damage [9]. Of note, ECFCs are negatively affected by adverse environments, and altered levels and or functions of ECFCs have been linked to CV events [17, 22, 42, 43].
Machine learning models. Performance of validated machine learning models to discriminate between ECFCs treated with either the serum from: a) critical patients; b) from asymptomatic PCR+ donors samples; c) from asymptomatic IgG + donors samples; and d) from COVID negative donors
| Model | Acc | AUC | TP-FP | Recall | Kappa | Gene names |
|---|---|---|---|---|---|---|
| MLR | 1.00 | 1.00 | 1.00-0.00 | 1.00 | 1.00 | SDCBP, CAPN5, HTRA2, STX10, ALDH1A1, MT-ND1, RIN1 |
| NB | 0.88 | 0.95 | 0.88-0.04 | 0.88 | 0.83 | C5ORF51, MCRIP1, CCS, EIF3H, RPP40, CD2BP2, ALDH1A1, MT-ND1, COX7C, HNRNPUL2, LSM12, CCNYL1 |
| LSVM | 0.97 | 0.98 | 0.97-0.01 | 0.97 | 0.96 | DDX39A, SDCBP, CAPN5, HTRA2, STX10, ALDH1A1, APOH, MT-ND1, RIN1 |
Acc: accuracy, AUC: area under the receiver operating characteristic curve, TP: true positive, FP: false, MLR: multinomial logistic regression, NB: Naïve Bayes, LSVM: linear support vector machines.
Proteomic changes in ECFCs related with cardiovascular-related pathologies. A) Altered functions and diseases related with CVDs in ECFCs incubated with the serum from critical patients (Crit vs Neg), provided by IPA. B) Functional network of CV related pathologies (Crit vs Neg), with proteins up- (red) or down-regulated (green). C) Altered diseases related with CVDs in ECFCs incubated with serum from asymptomatics (PCR+ and IgG+), compared with ECFCs+Neg samples. D) VCAM-1 levels increased in ECFCs in response to the serums from asymptomatic and critical donors compared with ECFCs+Neg samples. E) Representative images of the reticular structures formed by ECFCs after 24h incubation with the serum of Negative, asymptomatics and critical patients. Differences between ECFC+PCR and ECFC+Neg were statistically significant (*p < 0.05).
In-silico analysis with the potential interactions between altered proteins in the COVID-19 patients' serum and ECFCs. Some of the most relevant protein interactions connected to CVD have been highlighted.
Remarkably, our results indicate that the incubation of ECFCs with the serum of COVID-19 positive individuals, asymptomatic (PCR+ or IgG+) or critical patients, promotes changes at the protein level that resemble alterations associated endothelial dysfunction and viral infection. Indeed, different proteins associated to viral infection like the Long-chain Acyl-CoA synthetase 1 (ACSL1), Calpain-5 (CAPN5) or Syntaxin 10 (STX10), appeared down-regulated in ECFCs after stimulation with the serum of COVID-19 positive individuals. The last two, CAPN5 and STX10, were identified by predictive tools as highly discriminating proteins between ECFCs treated with “infected” serums and ECFCs incubated with COVID-19 negative serums (ECFCs+Neg). Noteworthy, ACSL1 has been proposed as an antiviral agent, by enhancing the production of interferon I (IFN-I) and mediating apoptosis through the PI3K/Akt signaling pathway in response to the avian leukosis virus [44]. ACSL1 down-regulation might reflect a viral strategy to reduce IFN-I levels, in agreement with previous studies demonstrating the potential of SARS-CoV-2 to prevent IFN-I signaling, and also to down-regulate the sensitivity and response of SARS-CoV-2 to IFN-I [45, 46]. On the other hand, CPN5 down-regulation might represent a protective mechanism in ECFCs, given the role that CPN5 and other calpain proteins seem to play in viral entry and replication [47-49]. Indeed, several calpain inhibitors such as MG132, II and XII, appear to inhibit SARS-CoV and SARS-CoV-2 at the early stages of viral replication [50, 51]. Finally STX10, a protein that facilitates vesicle´s fusion during intracellular trafficking of proteins and other cellular components [52], has been found to interact with the SARS-CoV-2 accessory proteins ORF3 and ORF7b, supporting its own replication and survival [46]. Thus, the modulation of STX10 or similar trafficking proteins might represent an alternative therapeutic target against this virus.
We also found protein changes only in ECFCs treated with the serum of asymptomatic individuals (PCR+ or IgG+), in agreement with previous results [7]. These changes correlated, among others, with mRNA surveillance or RNA transport, phagosomes and autophagy. Different studies have evaluated the potential role of autophagy into viral invasion and replication [53, 54]. Although autophagy represents a protective strategy from the host cells to eliminate the intruding viruses [55], viruses can interfere or evade the autophagic process, and even use the autophagic machinery for their own replication [56]. Recent reports indicated that SARS-CoV-2 efficiently avoids the anti-viral functions of autophagy, regulating autophagy by interaction of its factors ORF3 and NSP6 with cell host autophagic factors such as WIPI2 or LAMP2 proteins [57], two proteins altered in response to the serums from COVID-19 patients.
Remarkably, the serum of critical patients promoted the highest number of protein changes in ECFCs, with a protein profile clearly differing from the rest of the conditions analyzed. Among them, Galectins-8 and 9 (LGALS8 and LGALS9) appeared up-regulated. LGALS9 has been associated to the severity of HIV viral infection [58, 59], and its expression increases in response to many different viruses, including HIV, HCV, hepatitis B virus, herpes simplex virus, influenza virus, or dengue virus [60]. Similarly, LGALS8 appears to recognize the SARS-CoV-2 protein spike S1, highly glycosylated, which activates an antiviral autophagy mechanism, which SARS-CoV-2 counteracts by cleavage of LGALS8 [61]. Therefore, LGALS8 up-regulation might represent a protective mechanism against the virus that is triggered only under severe conditions (ECFCs+Crit). Further studies are needed to validate this hypothesis.
Like LGALS8, Polypyrimidine tract binding protein (PTBP1) and two members of the lysosomal cysteine protease family, Cathepsin L (CTSL) and D (CTSD), were also highly upregulated in ECFCs+Crit cells. PTBP1 binds mRNA and is essential for viral translation and replication [62]. Recent studies suggest that PTBP1 cleavage or inhibition might promote an inhibitory effect on SARS-CoV-2 replication [61]. Similarly, CTSL, a matrix-degrading enzyme upregulated in chronic inflammation [63], is used by SARS-CoV and SARS-CoV-2 viruses to cleave and activate the spike protein S between the residues Thr696 and Met697 in the S1-S2 domains, promoting the S-protein mediated cell-cell fusion and the release of the virus´s genome into the host cell[64, 65], Indeed, several CTSL inhibitors have shown promising results by impairing the entry of the virus and further replication [64, 65].
Several proteins associated with mitochondrial dysfunction, such as Cytochrome c oxidase subunit 7C (COX7C) and NADH-ubiquinone oxidoreductase chain 1 (MT-ND1), were also highlighted by machine learning algorithms. Both proteins were downregulated in ECFCs+PCR and more significantly in ECFCs+Crit. Notably, like many other viruses, SARS-CoV-2 is thought to modulate mitochondrial dynamics in its own benefit [66-68], by sending its genetic material towards the mitochondria to influence ROS production, mitophagy, iron storage, platelet coagulability, and cytokine production stimulation, supporting viral replication [69, 70]. Thus, the infection of endothelial cells by SARS-COV-2 might contribute to mitochondrial dysfunction and oxidative stress, key players on the initiation of chronic inflammation and endothelial damage [71]. Drugs targeting mitochondria and/or some of the proteins highlighted here could be considered as potential tools for protecting the endothelium in severe forms of COVID-19 [72].
Amidst all, many protein changes in ECFCs+Crit cells were linked to CVDs, including cardiomyopathy (also seen in ECFCs+PCR cells), ventricular dysfunction, vaso-occlusion and thrombosis, ischemic stroke or even kidney thrombosis. In agreement with these results, functional assays reported that the levels of VCAM1 were up-regulated in response to all positive serums, as already seen in COVID-19 patients [73-75], while the angiogenic potential of ECFCs was impaired. Both situations are indicative of endothelial dysfunction associated with cardiovascular events [33, 75].
Some of the CVDs-related proteins identified were connected, by in silico analysis, to several proteins that have been found altered in the serum of critical patients (Figure 7). For example, the Platelet factor 4 (PF4), a protein highly up-regulated only in ECFCs+Crit cells, was connected to PPBP, THBS1, FN1, CRP or vWF. Some of these proteins, including PF4, participate in the coagulation pathway, which becomes highly activated in severe COVID-19 patients, being responsible of thrombotic events [76, 77]. For instance, PF4 interaction with heparin promotes platelet aggregation and thrombi formation. Moreover, the formation of PF4-vWF complexes might propagate the risk of thrombosis in an heparin dependent manner [78]. PF4 is also highly secreted in response to viral infection, contributing to neutrophils recruitment, among others [79]. Finally, high levels of anti-PF4/heparin antibodies have been found in hospitalized patients, although this seem to be associated with the severity of COVID-19 rather than a marker of thrombotic risk [80]. Thus, up-regulation of serum markers such as vWF or CRP might be directly or indirectly associated with the upregulation of PF4 or other thrombotic markers in ECFCs.
LAMP2 or SURF4 were also up-regulated in ECFCs in response to the serum of critical patients as well as in ECFCs with PCR+ serum (at the highest peak of infection) and, interestingly, in individuals that had overcome the infection without apparent symptoms (PCR-/IgG+). These proteins were connected in silico to several serum markers upregulated in critical COVID-19 patients, including the chaperone Heat shock protein HSPA5, also called glucose regulating protein 78 (GRP78), a glycoprotein upregulated as result of ER stress mediated by COVID-19 infection [81]. Regarding LAMP2, this protein has been associated to autophagy, more precisely, chaperone mediated autophagy (CMA), activated against oxygen and glucose deprivation, conferring cardiomyocyte resistance against such stress in vitro [82]. Moreover, LAMP2 deficiency and disrupted autophagy are responsible of Danon disease, a rare cardiomyopathy that usually leads to profound hypertrophic cardiomyopathy resulting in death or requiring transplantation in men [83, 84]. On the other hand, nothing has been described, to our knowledge, associating directly LAMP2 with SARS-CoV-2 infection. Given the implication of LAMP2 in autophagy and the effect that impaired autophagy has over the CV system, future studies should evaluate the potential modulation of this virus over LAMP2 as well as other autophagic proteins as an alternative to block or avoid the progression towards more complicated and long-term vascular situations.
Finally, the cargo receptor SURF4 is an integral ER membrane protein involved in the assembly and packaging of proteins into ER‐derived transport vesicles. This protein regulates, among others, insulin [85] or PKSL9[86] secretion via ER export. Since both proteins are important regulators of both glucose and plasma cholesterol levels, up-regulation of SURF4 in ECFCs+Crit might reflect, again, the capacity of SARS-CoV-2 to alter lipid and glucose metabolism [87] in its own benefit.
Conclusion
Overall, the incubation of adult ECFCs with the serum factors of SARS-CoV-2 infected individuals constitutes an optimal approach to evaluate the endothelial cells response to SARS-CoV-2 depending on the severity of COVID-19 disease, in agreement with previous results [7]. Indeed, machine learning algorithms have reported some specific proteins such as STX10, SDCBP, CAPN5, MT-ND1 or ALDH1A1 as highly discriminating proteins between the groups compared. Many proteins identified here have been associated to viral invasion, extravasation and replication, while many others provide insights of the potential mechanisms of the virus to alter the cell host machinery in its own benefit (autophagy, mitochondrial dysfunction, etc). Moreover, the serum factors of infected individuals compromised the angiogenic potential of ECFCs, while promoted changes in the endothelial cells resembling cardiovascular-related pathologies. These changes might as well explain the activation of long-term vascular sequelae after SARS-CoV-2 infection.
While future research is required to determine which factors might indeed be promoting the protein changes seen here, as well as to further validate the involvement of the proteins identified, some of the proteins highlighted might be taken as potential candidates for therapeutic approaches to neutralize SARS-CoV-2 effect over the endothelium and hopefully to prevent potential CV events in COVID patients and post-COVID individuals.
Abbreviations
Area under the ROC curve (AUC); C-reactive protein (CRP); Calpain-5 (CAPN5); Cardiovascular (CV); Cardiovascular diseases (CVDs); Cathepsin L (CTSL); Cathepsin D (CTSD); Circulating angiogenic cells (CACs); Circulating endothelial cells (CECs); Creatine kinase (CK); Cytochrome C oxidase subunit 7C (COX7C); Dengue virus (DENV); Endothelial colony forming cells (ECFCs); Galectin-8 (LGALS8); Galectin-9 (LGALS9); Hepatitis B virus (HBV); Herpes simplex virus (HSV); Human immunodeficiency virus (HIV); Interferon I (IFN-I); Label free quantitative (LFQ); Linear support vector machines (LSVM); Long-chain Acyl-CoA synthetase 1 (ACSL1); Lysosome-associated membrane protein 2 (LAMP2); Mass spectrometry (MS); Multinomial logistic regression (MLR); N-terminal (NT); NADH-ubiquinone oxidoreductase chain 1 (MT-ND1); Näive Bayes (NB); Platelet factor 4 (PF4); Polypyrimidine tract binding protein (PTBP1); Principal component analysis (PCA); proB-type natriuretic peptide (BNP); procalcitonin (PCT); Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); Surfeit locus protein 4 (SURF4); Syntaxin 10 (STX10).
Supplementary Material
Supplementary tables.
Acknowledgements
Thanks to the Andalusian Bioinformatics Platform Center, Malaga University for the assistance with IPA software. Some images were obtained via SMART (https://smart.servier.com). Thanks to Dr.Vila-del Sol and A. Marquina-Rodriguez at the Flow Cytometry Core Facility of the National Paraplejic Hospital, Toledo; and also to the Biomedical Core Facility (SCIBM), Cadiz University, where part of the assays were carried out.
Funding
This study was supported by GLOBALCAJA-Ayuda COVID-19; and Fondo Supera COVID-19, funded by Banco Santander and CRUE universidades, Ref. IPSA-COVID-19, and the Institute of Health Carlos III, ISCIII (PI18-00427, PI20-00716), co-funded by European Regional Development “A way to make Europe”.
Ethics approval and consent to participate
The study was approved by the local Ethics Committee, in accordance to Spanish and European Union Regulations and it follows the principles outlined in the Declaration of Helsinki. All donors provided written informed consent prior to sample collection.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [88] partner repository with the data set identifier PXD034620.
Author contributions
MMT, MRI, MDN and MRI participated in patient recruitment and determination of patient infection by RT-q-PCR. RML designed and managed the logistics of recruitment, collection, stratification and samples storage. LBC, MRT and SEA performed ELISA assays to confirm the patients' infective stage (IgG/IgM). RML, performed the extraction and characterization of ECFCs, while LBC and ISG performed in vitro assays. LBC and SB performed the proteomic analysis and functional/biological analyses. DSM performed machine learning classification. LBC, SDB, DSM and MCD designed figures and tables. LBC and MCD wrote the main draft. LBC, MCD, SDB, MRL, DSM, JAM, EB and RML evaluated the final data, edited and revised the manuscript. RML and MCD conceptualized the project and got the funding. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Snell J. SARS-CoV-2 infection and its association with thrombosis and ischemic stroke: a review. Am J Emerg Med. 2021;40:188-92
2. Xie Y, Xu E, Bowe B, Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat Med. 2022;28:583-90
3. Pons S, Fodil S, Azoulay E, Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit Care. 2020;24:353
4. Ma Z, Yang KY, Huang Y, Lui KO. Endothelial contribution to COVID-19: an update on mechanisms and therapeutic implications. J Mol Cell Cardiol. 2022;164:69-82
5. Evans PC, Rainger GE, Mason JC, Guzik TJ, Osto E, Stamataki Z. et al. Endothelial dysfunction in COVID-19: a position paper of the ESC Working Group for Atherosclerosis and Vascular Biology, and the ESC Council of Basic Cardiovascular Science. Cardiovasc Res. 2020;116:2177-84
6. Haffke M, Freitag H, Rudolf G, Seifert M, Doehner W, Scherbakov N. et al. Endothelial dysfunction and altered endothelial biomarkers in patients with post-COVID-19 syndrome and chronic fatigue syndrome (ME/CFS). J Transl Med. 2022;20:138
7. Beltran-Camacho L, Eslava-Alcon S, Rojas-Torres M, Sanchez-Morillo D, Martinez-Nicolas MP, Martin-Bermejo V. et al. The serum of COVID-19 asymptomatic patients up-regulates proteins related to endothelial dysfunction and viral response in circulating angiogenic cells ex-vivo. Mol Med. 2022;28:40
8. Chioh FW, Fong SW, Young BE, Wu KX, Siau A, Krishnan S. et al. Convalescent COVID-19 patients are susceptible to endothelial dysfunction due to persistent immune activation. Elife. 2021 10
9. Poyatos P, Luque N, Eizaguirre S, Sabater G, Sebastian L, Francisco-Albesa I. et al. Post-COVID-19 patients show an increased endothelial progenitor cell production. Transl Res. 2022;243:14-20
10. Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270-5
11. Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M. et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905-13 e7
12. Zhang M, Wang P, Luo R, Wang Y, Li Z, Guo Y. et al. Biomimetic Human Disease Model of SARS-CoV-2 Induced Lung Injury and Immune Responses on Organ Chip System. Adv Sci (Weinh). 2020: 2002928.
13. Liu F, Han K, Blair R, Kenst K, Qin Z, Upcin B. et al. SARS-CoV-2 Infects Endothelial Cells In vivo and In vitro. Front Cell Infect Microbiol. 2021;11:701278
14. Hur J, Yoon CH, Kim HS, Choi JH, Kang HJ, Hwang KK. et al. Characterization of two types of endothelial progenitor cells and their different contributions to neovasculogenesis. Arterioscler Thromb Vasc Biol. 2004;24:288-93
15. Medina RJ, Barber CL, Sabatier F, Dignat-George F, Melero-Martin JM, Khosrotehrani K. et al. Endothelial Progenitors: A Consensus Statement on Nomenclature. Stem Cells Transl Med. 2017;6:1316-20
16. Alphonse RS, Vadivel A, Fung M, Shelley WC, Critser PJ, Ionescu L. et al. Existence, functional impairment, and lung repair potential of endothelial colony-forming cells in oxygen-induced arrested alveolar growth. Circulation. 2014;129:2144-57
17. Basile DP, Yoder MC. Circulating and tissue resident endothelial progenitor cells. J Cell Physiol. 2014;229:10-6
18. Lin RZ, Moreno-Luna R, Munoz-Hernandez R, Li D, Jaminet SC, Greene AK. et al. Human white adipose tissue vasculature contains endothelial colony-forming cells with robust in vivo vasculogenic potential. Angiogenesis. 2013;16:735-44
19. Sargento-Freitas J, Aday S, Nunes C, Cordeiro M, Gouveia A, Silva F. et al. Endothelial Progenitor Cells influence acute and subacute stroke hemodynamics. J Neurol Sci. 2018;385:119-25
20. Oliveras A, Soler MJ, Martinez-Estrada OM, Vazquez S, Marco-Feliu D, Vila JS. et al. Endothelial progenitor cells are reduced in refractory hypertension. J Hum Hypertens. 2008;22:183-90
21. Tan CMJ, Lewandowski AJ, Williamson W, Huckstep OJ, Yu GZ, Fischer R. et al. Proteomic Signature of Dysfunctional Circulating Endothelial Colony-Forming Cells of Young Adults. J Am Heart Assoc. 2021;10:e021119
22. Fadini GP, Mehta A, Dhindsa DS, Bonora BM, Sreejit G, Nagareddy P. et al. Circulating stem cells and cardiovascular outcomes: from basic science to the clinic. Eur Heart J. 2020;41:4271-82
23. Palmisano G, Lendal SE, Engholm-Keller K, Leth-Larsen R, Parker BL, Larsen MR. Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat Protoc. 2010;5:1974-82
24. Meier F, Brunner AD, Frank M, Ha A, Bludau I, Voytik E. et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat Methods. 2020;17:1229-36
25. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731-40
26. Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007-2015.
27. Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Research. 2021;49:W388-W96
28. Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P. et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data. 2017;4:170151
29. Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507-17
30. Frank E, Hall M, Witten I. The Weka Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”. 2016
31. Eslava-Alcon S, Extremera-Garcia MJ, Sanchez-Gomar I, Beltran-Camacho L, Rosal-Vela A, Munoz J. et al. Atherosclerotic Pre-Conditioning Affects the Paracrine Role of Circulating Angiogenic Cells Ex-Vivo. Int J Mol Sci. 2020 21
32. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523
33. Troncoso MF, Ortiz-Quintero J, Garrido-Moreno V, Sanhueza-Olivares F, Guerrero-Moncayo A, Chiong M. et al. VCAM-1 as a predictor biomarker in cardiovascular disease. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166170
34. Huang I, Pranata R, Lim MA, Oehadian A, Alisjahbana B. C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis. 2020;14:1753466620937175
35. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med. 2020;383:2255-73
36. Shafi AMA, Shaikh SA, Shirke MM, Iddawela S, Harky A. Cardiac manifestations in COVID-19 patients-A systematic review. J Card Surg. 2020;35:1988-2008
37. Guzik TJ, Mohiddin SA, Dimarco A, Patel V, Savvatis K, Marelli-Berg FM. et al. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020;116:1666-87
38. Calderon-Dominguez M, Trejo-Gutierrez E, Gonzalez-Rovira A, Beltran-Camacho L, Rojas-Torres M, Eslava-Alcon S. et al. Serum microRNAs targeting ACE2 and RAB14 genes distinguish asymptomatic from critical COVID-19 patients. Mol Ther Nucleic Acids. 2022;29:76-87
39. Gutmann C, Khamina K, Theofilatos K, Diendorfer AB, Burnap SA, Nabeebaccus A. et al. Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc Res. 2022;118:461-74
40. Mejia-Renteria H, Travieso A, Sagir A, Martinez-Gomez E, Carrascosa-Granada A, Toya T. et al. In-vivo evidence of systemic endothelial vascular dysfunction in COVID-19. Int J Cardiol. 2021;345:153-5
41. Lampsas S, Tsaplaris P, Pantelidis P, Oikonomou E, Marinos G, Charalambous G. et al. The Role of Endothelial Related Circulating Biomarkers in COVID-19. A Systematic Review and Meta-analysis. Curr Med Chem. 2021
42. Bertagnolli M, Xie LF, Paquette K, He Y, Cloutier A, Fernandes RO. et al. Endothelial Colony-Forming Cells in Young Adults Born Preterm: A Novel Link Between Neonatal Complications and Adult Risks for Cardiovascular Disease. J Am Heart Assoc. 2018 7
43. Nafisa A, Gray SG, Cao Y, Wang T, Xu S, Wattoo FH. et al. Endothelial function and dysfunction: Impact of metformin. Pharmacol Ther. 2018;192:150-62
44. Zhang Q, Xie T, Mo G, Zhang Z, Lin L, Zhang X. ACSL1 Inhibits ALV-J Replication by IFN- Signaling and PI3K/Akt Pathway. Front Immunol. 2021;12:774323
45. Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M. et al. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020 94
46. Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C. et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246-52
47. Ono Y, Saido TC, Sorimachi H. Calpain research for drug discovery: challenges and potential. Nat Rev Drug Discov. 2016;15:854-76
48. Bruening J, Weigel B, Gerold G. The Role of Type III Interferons in Hepatitis C Virus Infection and Therapy. J Immunol Res. 2017;2017:7232361
49. Barnard DL, Hubbard VD, Burton J, Smee DF, Morrey JD, Otto MJ. et al. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-D-N4-hydroxycytidine. Antivir Chem Chemother. 2004;15:15-22
50. Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T. et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678-92
51. Schneider M, Ackermann K, Stuart M, Wex C, Protzer U, Schatzl HM. et al. Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. J Virol. 2012;86:10112-22
52. Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180:159-72
53. Delorme-Axford E, Klionsky DJ. Highlights in the fight against COVID-19: does autophagy play a role in SARS-CoV-2 infection? Autophagy. 2020;16:2123-7
54. Pereira G, Leao A, Erustes AG, Morais IBM, Vrechi TAM, Zamarioli LDS. et al. Pharmacological Modulators of Autophagy as a Potential Strategy for the Treatment of COVID-19. Int J Mol Sci. 2021 22
55. Koepke L, Hirschenberger M, Hayn M, Kirchhoff F, Sparrer KM. Manipulation of autophagy by SARS-CoV-2 proteins. Autophagy. 2021;17:2659-61
56. Carmona-Gutierrez D, Bauer MA, Zimmermann A, Kainz K, Hofer SJ, Kroemer G. et al. Digesting the crisis: autophagy and coronaviruses. Microb Cell. 2020;7:119-28
57. Miao G, Zhao H, Li Y, Ji M, Chen Y, Shi Y. et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56:427-42 e5
58. Abdel-Mohsen M, Chavez L, Tandon R, Chew GM, Deng X, Danesh A. et al. Human Galectin-9 Is a Potent Mediator of HIV Transcription and Reactivation. PLoS Pathog. 2016;12:e1005677
59. Golden-Mason L, Rosen HR. Galectin-9: Diverse roles in hepatic immune homeostasis and inflammation. Hepatology. 2017;66:271-9
60. Lai JH, Luo SF, Wang MY, Ho LJ. Translational Implication of Galectin-9 in the Pathogenesis and Treatment of Viral Infection. Int J Mol Sci. 2017 18
61. Pablos I, Machado Y, de Jesus HCR, Mohamud Y, Kappelhoff R, Lindskog C. et al. Mechanistic insights into COVID-19 by global analysis of the SARS-CoV-2 3CL(pro) substrate degradome. Cell Rep. 2021;37:109892
62. Florez PM, Sessions OM, Wagner EJ, Gromeier M, Garcia-Blanco MA. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J Virol. 2005;79:6172-9
63. Cao Y, Liu X, Li Y, Lu Y, Zhong H, Jiang W. et al. Cathepsin L activity correlates with proteinuria in chronic kidney disease in humans. Int Urol Nephrol. 2017;49:1409-17
64. Zhao MM, Yang WL, Yang FY, Zhang L, Huang WJ, Hou W. et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther. 2021;6:134
65. Gomes CP, Fernandes DE, Casimiro F, da Mata GF, Passos MT, Varela P. et al. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front Cell Infect Microbiol. 2020;10:589505
66. Khan M, Syed GH, Kim SJ, Siddiqui A. Mitochondrial dynamics and viral infections: A close nexus. Biochim Biophys Acta. 2015;1853:2822-33
67. Anand SK, Tikoo SK. Viruses as modulators of mitochondrial functions. Adv Virol. 2013;2013:738794
68. Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA. et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179-86
69. de Las Heras N, Martin Gimenez VM, Ferder L, Manucha W, Lahera V. Implications of Oxidative Stress and Potential Role of Mitochondrial Dysfunction in COVID-19: Therapeutic Effects of Vitamin D. Antioxidants (Basel). 2020 9
70. Ganji R, Reddy PH. Impact of COVID-19 on Mitochondrial-Based Immunity in Aging and Age-Related Diseases. Frontiers in Aging Neuroscience. 2021 12
71. Pelisek J, Reutersberg B, Greber UF, Zimmermann A. Vascular dysfunction in COVID-19 patients: update on SARS-CoV-2 infection of endothelial cells and the role of long non-coding RNAs. Clin Sci (Lond). 2022;136:1571-90
72. Chernyak BV, Popova EN, Zinovkina LA, Lyamzaev KG, Zinovkin RA. Mitochondria as Targets for Endothelial Protection in COVID-19. Frontiers in Physiology. 2020 11
73. Bruni F, Charitos P, Lampart M, Moser S, Siegemund M, Bingisser R. et al. Complement and endothelial cell activation in COVID-19 patients compared to controls with suspected SARS-CoV-2 infection: A prospective cohort study. Front Immunol. 2022;13:941742
74. Tong M, Jiang Y, Xia D, Xiong Y, Zheng Q, Chen F. et al. Elevated Expression of Serum Endothelial Cell Adhesion Molecules in COVID-19 Patients. J Infect Dis. 2020;222:894-8
75. Birnhuber A, Fließer E, Gorkiewicz G, Zacharias M, Seeliger B, David S. et al. Between inflammation and thrombosis: endothelial cells in COVID-19. European Respiratory Journal. 2021;58:2100377
76. Cai Z, Greene MI, Zhu Z, Zhang H. Structural Features and PF4 Functions that Occur in Heparin-Induced Thrombocytopenia (HIT) Complicated by COVID-19. Antibodies (Basel). 2020 9
77. Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7:e438-e40
78. Johnston I, Sarkar A, Hayes V, Koma GT, Arepally GM, Chen J. et al. Recognition of PF4-VWF complexes by heparin-induced thrombocytopenia antibodies contributes to thrombus propagation. Blood. 2020;135:1270-80
79. Wilkinson JM, Ladinig A, Bao H, Kommadath A, Stothard P, Lunney JK. et al. Differences in Whole Blood Gene Expression Associated with Infection Time-Course and Extent of Fetal Mortality in a Reproductive Model of Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Infection. PLoS One. 2016;11:e0153615
80. Pascreau T, Ballester MC, Van Dreden P, Zia-Chahabi S, Zuber B, Choucair J. et al. The high frequency of anti-PF4/heparin antibodies in patients with COVID-19 is neither related to heparin treatment or to an increased incidence of thrombosis. Clin Chem Lab Med. 2021;59:e405-e8
81. Fu J, Wei C, He J, Zhang L, Zhou J, Balaji KS. et al. Evaluation and characterization of HSPA5 (GRP78) expression profiles in normal individuals and cancer patients with COVID-19. Int J Biol Sci. 2021;17:897-910
82. Cui L, Zhao LP, Ye JY, Yang L, Huang Y, Jiang XP. et al. The Lysosomal Membrane Protein Lamp2 Alleviates Lysosomal Cell Death by Promoting Autophagic Flux in Ischemic Cardiomyocytes. Front Cell Dev Biol. 2020;8:31
83. Endo Y, Furuta A, Nishino I. Danon disease: a phenotypic expression of LAMP-2 deficiency. Acta Neuropathol. 2015;129:391-8
84. Rowland TJ, Sweet ME, Mestroni L, Taylor MR. Danon disease - dysregulation of autophagy in a multisystem disorder with cardiomyopathy. J Cell Sci. 2016;129:2135-43
85. Saegusa K, Matsunaga K, Maeda M, Saito K, Izumi T, Sato K. Cargo receptor Surf4 regulates endoplasmic reticulum export of proinsulin in pancreatic beta-cells. Commun Biol. 2022;5:458
86. Emmer BT, Hesketh GG, Kotnik E, Tang VT, Lascuna PJ, Xiang J. et al. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife. 2018 7
87. Keihanian F, Bigdelu L. Cardiovascular Considerations in COVID19: A Comprehensive Review. Ther Clin Risk Manag. 2020;16:1089-97
88. Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442-D50
Author contact
![]() Corresponding authors: Rafael Moreno-Luna, PhD; rmlunajccm.es; Laboratorio de Neuroinflamación, i2-06, Hospital Nacional de Parapléjicos-SESCAM. CP 45071, Toledo, Spain; Mª Carmen Durán-Ruiz, PhD; maricarmen.duranuca.es; Biomedicine, Biotechnology and Public Health Department, Science Faculty, Cádiz University. Torre Sur. Avda. República Saharaui S/N, Polígono Río San Pedro, Puerto Real, 11519 Cádiz, Spain.
Corresponding authors: Rafael Moreno-Luna, PhD; rmlunajccm.es; Laboratorio de Neuroinflamación, i2-06, Hospital Nacional de Parapléjicos-SESCAM. CP 45071, Toledo, Spain; Mª Carmen Durán-Ruiz, PhD; maricarmen.duranuca.es; Biomedicine, Biotechnology and Public Health Department, Science Faculty, Cádiz University. Torre Sur. Avda. República Saharaui S/N, Polígono Río San Pedro, Puerto Real, 11519 Cádiz, Spain.

 Global reach, higher impact
Global reach, higher impact