Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(6):1748-1763. doi:10.7150/ijbs.81000 This issue Cite
Review
Novel insights into the interplay between m6A modification and programmed cell death in cancer
Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, Nanjing, China.
*These authors contributed equally.
Received 2022-11-18; Accepted 2023-3-8; Published 2023-3-13
Abstract
N6-methyladenosine (m6A) methylation, the most prevalent and abundant RNA modification in eukaryotes, has recently become a hot research topic. Several studies have indicated that m6A modification is dysregulated during the progression of multiple diseases, especially in cancer development. Programmed cell death (PCD) is an active and orderly method of cell death in the development of organisms, including apoptosis, autophagy, pyroptosis, ferroptosis, and necroptosis. As the study of PCD has become increasingly profound, accumulating evidence has revealed the mutual regulation of m6A modification and PCD, and their interaction can further influence the sensitivity of cancer treatment. In this review, we summarize the recent advances in m6A modification and PCD in terms of their interplay and potential mechanisms, as well as cancer therapeutic resistance. Our study provides promising insights and future directions for the examination and treatment of cancers.
Keywords: N6-methyladenosine, cancer, programmed cell death, apoptosis, autophagy, pyroptosis, ferroptosis, necroptosis
Introduction
Studies have revealed that more than 160 chemical modifications occur in RNA molecules, and m6A modification is the sixth N atom of adenine to be methylated (1). It is the most common post-transcriptional modification and exerts a variety of essential biological functions on mRNA and ncRNA (2, 3). M6A modification sites are often located in stop codons and the 3'-UTR region with a typical consensus sequence RRACH (R = G or A and H = A, C, or U) (4), and are regulated by writers, erasers, and readers. Studies have indicated that m6A modification is associated with many physiological processes, including the alternative splicing of pre-mRNAs, mRNA degradation, mRNA stabilization, miRNA processing, and cap-independent translation (5). Additionally, m6A plays an indispensable role in cancer and other diseases (6,7).
Programmed cell death (PCD) is an active and orderly process of cell death, including apoptosis, autophagy, ferroptosis, pyroptosis, and necroptosis (8). To balance the homeostasis of the internal environment, PCD can clear and maintain abnormal cells. In addition, aberrant PCD, which has been extensively manipulated to affect the development of cancer and other diverse diseases, has received increasing attention (9). Since the regulation between m6A and apoptosis was first elaborated, emerging studies have focused on abnormal m6A levels as key regulators of PCD. In this review, we introduce the complex role of m6A and the latest progress regarding the connection between m6A modifications and PCD in diverse diseases. Additionally, the future clinical applications of m6A-modified PCD in chemotherapies and precision medicine are also discussed.
M6A regulators
M6A modification is a dynamic and reversible progression that is modulated by methyltransferases and demethylases (10)(Fig.1). Regarding the M6A writers, such as methyltransferase-like protein 3/14 (METTL3/14), Wilms tumor-associated protein (WTAP), and vir-like m6A methyltransferase associated (VIRMA) (11) are the key proteins responsible for catalyzing m6A methylation on RNA (12). METTL3 and METTL14 are the catalytic cores of MTCs (13). Methyltransferase-like protein 3 (METTL3), an S-adenosylmethionine (SAM)-binding protein, catalyzes the transfer of methyl groups in SAM (14). Methyltransferase-like protein 14 (METTL14), another active component of MTC, is a support enzyme devoid of catalytic activity and can form a stable complex with METTL3 to stabilize the structure of MTC (15). Other subunits, such as Wilms tumor1-associated protein (WTAP) (16) and RNA binding motif protein 15 (RBM15) (17), can attract MTCs to specific regions of RNA to induce methylation. METTL3, METTL5 (18), and METTL16 (19), the newly discovered independent RNA methylases, as catalytic subunits, can also catalyze m6A modification via methyltransferase domains. Zinc finger CCCH domain-containing protein 13 (ZC3H13) enhances the MTC catalytic function by interacting with WTAP (20). Conversely, m6A erasers, such as fat mass and obesity-associated protein (FTO) and ALKB homolog 5 (ALKHB5), perform the function of demethylating m6A modified bases (21,22). ALKBH3, a newly discovered demethylase, can also participate in this process via a similar mechanism (23). Moreover, m6A readers, m6A binding proteins recognize m6A modifications, including YTH domain-containing family protein (YTHDF1-3 and YTHDC1-2) (24, 25, 26, 27), heterogeneous nuclear ribonucleoprotein (HNRNP) protein families (28), eukaryotic translation initiation factor 3 (eIF3) (29), and insulin-like growth factor-2 mRNA-binding proteins 1/2/3 (IGF2BP1/2/3) (30, 31, 32), and identify and bind to m6A marks directly to regulate downstream mRNA translation, decay, and stability.
M6A modifications in RNA metabolism
As noted above, m6A methylation elicits a wide range of efforts on mRNA processing, splicing, translation, and decay by m6A writer-complex component and m6A reader, as well as erasers that affect RNA metabolism to determine RNA fate and function (5). The importance of m6A as a post-transcriptional modification has been recognized, but the evolution, function and regulation of individual m6A sites remain largely unknown. M6A modification sites are broadly found in stop codons and the 3'-UTR regions with the consensus sequence RRACH (in which R represents A or G, and H represents A, C or U) (4). M6A methylation sites may appear with different functional consequences depending on their location. Concretely speaking, some research revealed that transcripts with m6A in the 5′UTR and/or CDS were related to energy metabolism, mitochondrial function and intracellular signals, while methylation of transcripts in the 3′UTR mostly code for proteins involved in pathways linked to more specific metabolic processes such as 'acetyl-CoA or glycerol biosynthesis' and 'positive regulation of protein dephosphorylation' (33).
M6A regulators. M6A is deposited by writers, removed by erasers, and recognized by readers. Regulatory functions of m 6 A modification in RNA splicing, processing, translation and degradation.
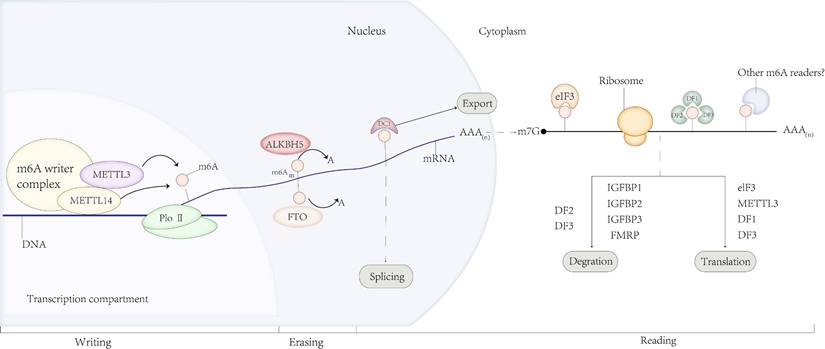
RNA metabolism to be more specific. For m6A modification in RNA processing, METTL3 labeled pri-miRNAs for recognition and processing by DGCR8 and promotes the initiation of miRNA biogenesis (34). Depletion of HNRNPA2B1 reduced the processing of primary microRNA-106b to enhance NSCLC cell growth (35). For m6A modification in RNA splicing, m6A modification prevents the essential splicing factor U2AF35 from recognizing the 3' splice site to inhibit RNA splicing (36). FTO regulates nuclear mRNA alternative splicing by binding with SRSF2 (37). YTHDC1 enhances an oncogenic RNA splicing of tumor suppressor RBM4 (38). For m6A modification in RNA degradation, YTHDF2 changes the localization of bound mRNA from the translatable pool to mRNA decay sites to regulate mRNA degradation (39). In colorectal cancer, YTHDF2 can also upregulate the mRNA stability of lncRNA STEAP3-AS1 to affect the development of cancer (40). METTL3 delays SOCS2 mRNA degradation to regulate liver cancer progression (41). For m6A modification in RNA translation, m6A in 3 'UTRs was found to improve translation efficiency by binding to YTHDF130(42), while m6A in 5' UTRs was reported to promote cap-independent translation under heat shock stress (43). M6A modifications can be found in mRNA and noncoding RNA (ncRNAs) to regulate gene expression in its 5′ or 3′ UTR. M6A modification not only drives translation on mRNA but also unexpected works on some ncRNAs (44, 45).
The double sword of m6A in human cancers
High-profile RNA epigenetic modification N6-methyladenosine (m6A), as a double-edged sword for cancer, can either promote or inhibit the occurrence and development of diverse diseases. However, the specific mechanism that determines the duality of m6A and the regulatory mechanism of m6A on core genes remain unclear. Here, we summarize the latest research progress on the intricate role of the same or similar function and different m6A-related enzymes in cancers.
The intricate role of different m6A-related enzymes in cancers
M6A methylation has been shown to play an important role in many cancer types, such as head and neck cancer (46), acute myeloid leukemia (AML) (47), glioblastoma (GBM) (48), nasopharyngeal carcinoma (NPC)(49), breast cancer (50), lung cancer (51), gastric cancer (52), pancreatic cancer (53), bladder cancer (54), hepatocellular cancer (HCC) (55), colorectal cancer (CRC)(56), endometrial cancer (57) etc (Fig.2). However, the role of m6A in different tumors is dynamic and contradictory.
WTAP can promote the progression and metastasis of NPC by increasing the stability of lncRNA DIAPH1-AS1 (58). METTL3 plays oncogenic roles in esophageal squamous cell carcinoma (ESCC) by decreasing the expression of APC, a tumor suppressor gene (59). In non-small cell lung cancer (NSCLC), METTL3 induces drug resistance and metastasis through the m6A-MALAT1-YAP axis (60). Similarly, in another common type of lung adenocarcinoma (LUAD), METTL3 increases the stability of lncRNA LCAT3, leading to the binding of FUBP1, activating the oncogenic molecule c-MYC, and promoting tumor proliferation, invasion, and metastasis (61). In addition, METTL3 can promote tumor development in LUAD by stimulating ENO1 translation mediated by YTHDF1 (62). Studies also revealed the critical part that m6A modification plays in drug resistance (63), (64). Recent studies have shown that m6A modification is important for immunoregulation. METTL3 depletion inhibits PD-L1 expression in an m6A-IGF2BP3-dependent manner, which in turn enhances antitumor immunity in breast cancer (65).
In GBM, the SPI1-induced downregulation of FTO promotes tumor progression by regulating pri-miR-10a processing in an m6A-dependent manner (66). In contrast, for NPM1-mutated AML, which accounts for approximately one-third of AML cases, FTO is aberrantly overexpressed and serves as a carcinogen by promoting the cell cycle and inhibiting apoptosis (67). Similarly, ALKBH5 promotes tumor growth and metastasis through the TRAF1-mediated activation of the NF-κB and MAPK signaling pathways in multiple myeloma (68).
To better understand the role of m6A in various diseases, it is important to examine methylated reading proteins that also play an integral role in tumor progression. YTHDF2 positively correlates with the grade and low prognosis of gliomas. YTHDF2 can activate NF-κB by accelerating the degradation of UBXN1 mRNA, thereby promoting glioma growth, proliferation, and metastasis (69). In addition, YTHDF1 promotes tumor progression in CRC by promoting ARHGEF2 translation and RhoA signaling (70). In AML, the m6A reader IGF2BP3 is associated with low prognosis and can accelerate the occurrence and development of tumors by increasing the stability of RCC2 (71).
In conclusion, m6A plays an important role in different tumors. However, the level of m6A in different tumors is bidirectional, both pro-cancer and anti-cancer, and the specific mechanism needs to be further explored.
The intricate role of m6A modification in different cancers.
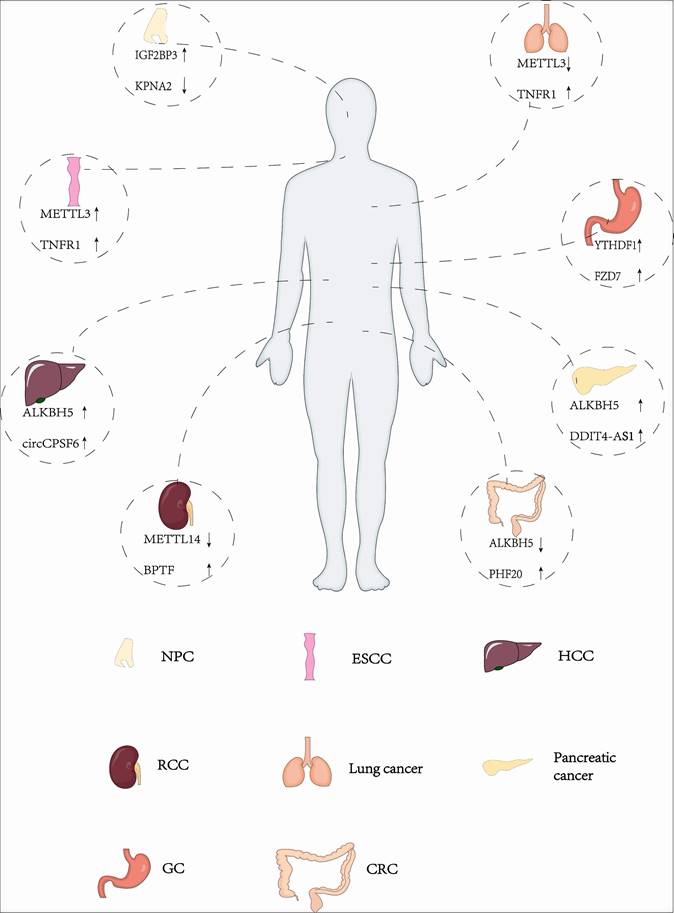
The contradictory role of m6A-related enzymes with the same or similar functions in cancers
Methylation-related enzymes with opposite effects may have contradictory expression, even in the same tumor. In gastric cancer, ALKBH5 is expressed at a low level and is strongly associated with clinical tumor distal metastasis and lymph node metastasis, while the silencing of ALKBH5 promotes tumor invasion and metastasis (72). In a study of gastric cancer, METTL3 was surprisingly under-expressed in the opposite direction and the overexpression of METTL3 inhibited gastric cancer progression through the methylation modification of circORC5 (73). In pancreatic cancer, ALKBH5 plays a pro-oncogenic role and ALKBH5 overexpression can promote tumor progression by downregulating potassium two-pore domain channel subfamily K member 15 and WISP2 antisense RNA 1 (KCNK15-AS1), an oncogene, to promote tumor growth invasion and metastasis (74). Conversely, METTL3, a methyltransferase, plays a pro-oncogenic role in pancreatic cancer. METTL3 accelerates the growth, invasion, and metastasis of pancreatic cancer by reducing SMS expression in an m6A-dependent manner (75).
Similarly, even m6A-related enzymes, which have the same roles in the same tumor, exhibit paradoxical roles. METTL14 is under-expressed in CRC and is associated with poor prognosis in patients. METTL14 inhibits tumor cell proliferation by abolishing the m6A level of XIST and augmenting XIST expression mediated by YTHDF2 (76). In another CRC study, METTL3 shows higher expression in CRC tissues than in normal tissues, and the overexpression of METTL3 promotes tumor progression by regulating the m6A-CRB3-Hippo axis (77). METTL3 induces GLUT1 translation to promote glucose uptake and lactate production, leading to the activation of mTROC1 signaling, thereby promoting tumor progression (78). In summary, it has been suggested that m6A plays conflicting roles not only in different tumors, but also in the same tumor. The specific reasons for this may include the heterogeneity of cell and tissue samples as well as the amount of m6A that varies dynamically in tumors. The same m6A-related enzymes may have other unknown potential roles. Different m6A-related proteins that perform the same function may have different unknown roles apart from their role in methylation, which can lead to opposite outcomes. The type of molecule regulated by m6A (pro-cancer or anti-cancer) may differ. The recognition of different reading proteins of m6A and the type of disease to a greater or lesser extent are also variable. The specific mechanisms also need to be further investigated.
The combination of m6A and PCD in cancers
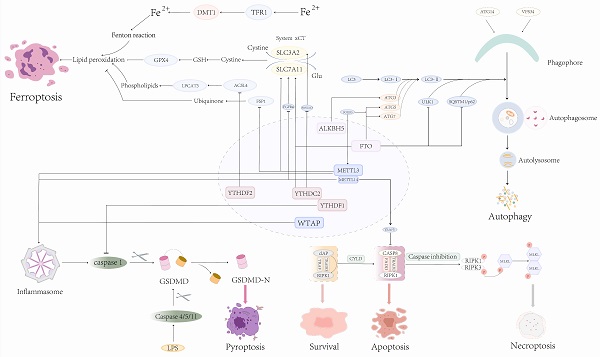
Here, we mainly discuss the connection between m6A and cell apoptosis, autophagy, pyroptosis, and ferroptosis, as well as necroptosis. M6A is controlled by regulatory factors (writers and erasers) and recognition factors (readers) to mediate downstream targets to regulate PCD (Fig. 3). It is certain that the connection of m6A and PCD pathways will provide new insights into the management of related diseases.
Role of m6A modification in mediating different types of PCD.
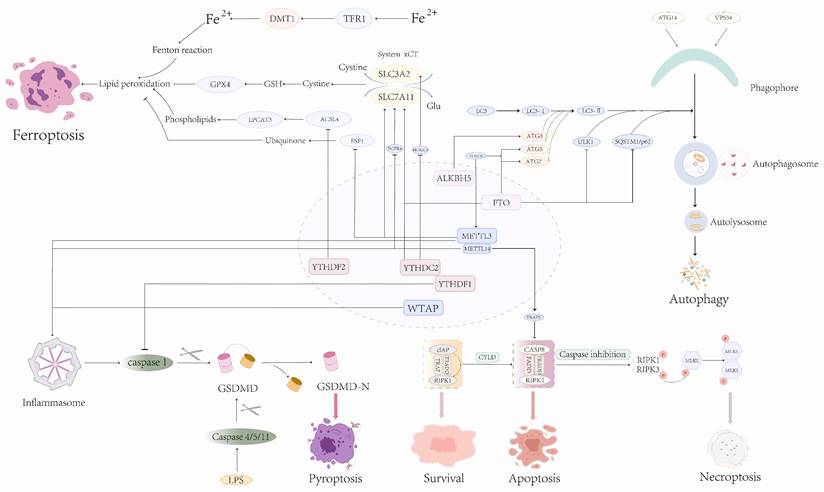
Role of m6A modification in mediating apoptosis.
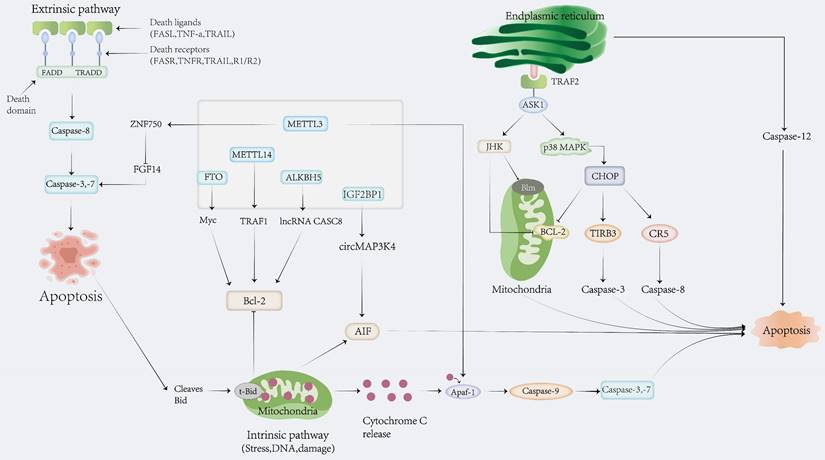
M6A and apoptosis
Programmed death refers to physiological death, which is a predetermined and tightly programmed cellular and molecular biological process in ontogeny. As one of the modes of programmed death, the apoptosis-related signaling pathway is classified into three types: classic mitochondria, endoplasmic reticulum, and exogenous death receptor pathways (79). Apoptosis-related molecular types include the Bcl-2 subfamily and the caspase family (80), (81). Since METTL3 depletion was reported to induce apoptosis and decrease AML development in 2017 (82), an increasing number of studies have revealed that apoptosis is regulated by m6A, which plays a crucial role in the occurrence and development of cancer by promoting or suppressing apoptosis (Fig.4).
In Alzheimer's disease, low levels of METTL3 lead to memory loss by promoting neuronal apoptosis with extensive synaptic loss, neuronal death, and multiple AD-related alterations, including oxidative stress and aberrant cell cycle events (83). The M6A-mediated upregulation of circMDK promotes cancer progression and apoptosis via the miR-346/874-3p-ATG16L1 axis (84). In the progression of osteosarcoma, the expression of ALKBH5 is low and its overexpression inhibits STAT3 activity to reduce cell proliferation and apoptosis in an m6A-YTHDF2-dependent manner (85). In contrast, ALKBH5 is upregulated in myeloma, and its inhibition represses the myeloma cell proliferation, invasion, and migration ability, while it promotes apoptosis (86).
Increasingly, studies have also demonstrated that the m6A-apoptosis axis plays a crucial role in the tumor microenvironment. The inhibition of IGF2BP1, as a crucial m6A reader protein, exerts a tumor suppressor effect in HCC by inducing apoptosis and subsequently activating immune cell infiltration as well as blocking PD-L1 expression to regulate the tumor immune microenvironment (87). COL10A1 secreted by cancer-associated fibroblasts (CAFs) and upregulated by METTL3, facilitates cell proliferation and represses apoptosis-induced oxidative stress in LUSC (88).
Apparently, m6A-apoptosis axis plays an indispensable role when it comes to addressing the problem of drug resistance in cancer. In a study of sunitinib resistance in renal cell carcinoma, TRAF1 increases significantly in sunitinib-resistant cells, while the TRAF1 overexpression promotes sunitinib resistance by modulating apoptosis in a METTL14-dependent manner (89). In gastric cancers, lncRNA ABL is significantly elevated, while the ABL overexpression inhibits GC cell apoptosis and enhances multidrug resistance. Mechanistically, ABL is stabilized by METTL3-mediated m6A modification and subsequently binds to APAF1 to block the apoptosome assembly and caspase-9/3 activation, thereby leading to increased sensitivity to chemotherapy (90). For chemoresistance in ESCC, the highly overexpressed ALKBH5-induced lncRNA CASC8 activate the Bcl2/Caspase3 pathway to decrease the cisplatin sensitivity of ESCC and promote tumor development (91).
M6A and autophagy
Autophagy is an endogenous defense process that relies on autophagic lysosomes that degrade their encapsulated contents to meet the metabolic needs of the cell and the renewal of some organelles, thus playing an important role in tumor development and evolution (92). It is characterized by the formation of autophagosomes (Frankelet al., 2017), autophagy-related genes (ATG), uncoordinated 51-like kinase 1 (ULK1), and transcription factor EB (TFEB), which act as important regulators of autophagy. In 2018, the first research on m6A and autophagy was conducted. FTO deficiency promotes the expression of ULK1, a key protein associated with autophagy, to delay tumor progression in an m6A-YTHDF2-dependent manner (93). Since then, valuable insights have been provided regarding the role of m6A-related autophagy in the occurrence and development of tumors.
Impaired autophagy has also been observed in the progression of osteoarthritis-synoviocytes. In terms of the mechanism, METTL3 decreases the expression of autophagy-related 7, an E-1 enzyme crucial for the formation of autophagosomes, enhances autophagic flux, and promotes cellular senescence and osteoarthritic progression (94). In the process of malignant skin transformation and tumorigenesis, FTO is upregulated and stabilized by low-level arsenic through the inhibition of p62-mediated selective autophagy (95). Similarly, FTO is also upregulated and closely related to autophagic flux in clear cell renal cell carcinoma, whereas FTO knockdown enhances autophagic flux and impairs tumor growth and metastasis (96). Methylated proteins can also regulate autophagy. YTHDF1 deficiency inhibits HCC autophagy, growth, and metastasis by promoting the translation of autophagy-related genes ATG2A and ATG14 in an m6A-dependent manner (97).
Emerging evidence indicates that autophagy regulated by m6A also influences the efficacy of immunotherapy. Melanoma tumorigenesis and anti-PD-1 resistance are promoted by m6A mRNA demethylase FTO, which is induced by metabolic starvation stress through the autophagy and NF-κB pathways (98). In addition, m6A methylation is involved in immune infiltration and autophagy in primary Sjögren's syndrome (pSS) (99). Furthermore, emerging studies have reported that the m6A-autophagy axis plays an important role in drug resistance. For gefitinib resistance in NSCLC cells, METTL3-mediated autophagy reverses this drug resistance by regulating β-elemene (100). The upregulated lncRNA ARHGAP5-AS1 is affected by autophagy, and SQSTM1 is responsible for transporting ARHGAP5-AS1 to autophagosomes in chemo-resistant gastric cancer cells (101). In addition, METTL3 improves the resistance of HCC cells to sorafenib by stabilizing FOXO3 mediated by YTHDF1 in an m6A-dependent manner, thereby inhibiting the expression of autophagy-related genes including ATG3, ATG5, ATG7, ATG12, and ATG16L1 (102).
In summary, the m6A-autophagy axis seems to be involved in the initiation and progression of different cancers and plays an important role in the immune phenotype and drug resistance. M6A-autophagy could be contributing to multifarious cancer progression and potentially represent a novel therapeutic target.
M6A and ferroptosis
Ferroptosis, a newly discovered type of programmed cell death, is an underlying therapeutic strategy for the inhibition of cancer occurrence and development (103). Since the discovery of ferroptosis in 2012 (104), an increasing number of studies have demonstrated the important role of ferroptosis in various cancers. Ferroptosis has been used to describe a highly complex process that requires the coordination of a series of signals from different organelles. The organelles involved include the endoplasmic reticulum, peroxisomes, and lysosomes. Several common mechanisms of ferroptosis are related to oxidative damage and antioxidant defense. Specifically, iron accumulates first, followed by lipid peroxidation, and finally the rupture of the cytoplasmic membrane occurs (105). Ferroptosis is an iron-dependent and non-apoptotic oxidative form of cell death, whose definitive hallmark genes are related to iron accretion and lipid peroxidation. It can be regulated at different levels, particularly in epigenetics. To date, an increasing number of studies have demonstrated a potential relationship between m6A modifications and ferroptosis (Table 1).
Doxorubicin, which plays a toxic role in the heart, upregulates METTL14 and promotes cardiomyocyte ferroptosis via the KCNQ1OT1-MIR-7-5P-TFRC axis. Therefore, targeting METTL14 and ferroptosis may provide a promising strategy for controlling DOX-induced cardiac injury (106). In addition, METTL3-mediated SLC7A11, a subunit of the Xc- system, enhances the ferroptotic resistance and promotes proliferation and apoptosis in LUAD (107). Similarly, m6A medicated SLC7A11 in ferroptosis has been found in hepatoblastoma (108), glioblastoma (109), thyroid cancer (110), and LUAD (111). For FSP1, an iron suppressor protein 1, another key factor in ferroptosis, miR-4443 is highly expressed in cisplatin-resistant tissue-derived exosomes in NSCLC, and miR-4443 overexpression can pass METLL3 by cisplatin treatment. This inhibits FSP1-mediated ferroptosis and promotes tumor growth (112).
In addition, m6A reading proteins have also been demonstrated to play an indispensable role in cancer. In LUAD, YTHDC2 is a ferroptosis inducer that promotes the development of cancer by targeting the SLC3A2 subunit of system Xc- (111).
Ferroptosis plays an important role in drug resistance. In recalcitrant HER2-positive breast cancer, FGFR4 knockdown reduces resistance to anti-HER2 therapy by activating ferroptosis. Mechanistically, FGFR4, modified by m6A, blocks glutathione synthesis and Fe2+ efflux efficiency via the β-catenin/TCF4-SLC7A11/FPN1 axis, resulting in excessive ROS production and labile iron pool accumulation (113). Hypoxia-induced lncRNA-CBSLR increases chemoresistance in gastric cancer by inhibiting ferroptosis. In detail, CBSLR decreases the stability of CBS mRNA in an m6A-YTHDF2-dependent manner, leading to the polyubiquitination and degradation of ACSL4, which decreases the pro-ferroptotic phosphatidylethanolamine (PE) (18:0/20:4) and PE (18:0/22:4) content to inhibit the activation of ferroptosis (114). In NSCLC, m6A-medicated miR-4443 also enhances cisplatin resistance through the inhibition of ferroptosis (112).
Although several ferroptosis-associated lncRNAs have been analyzed for their correlation with m6A-related genes in terms of immune efficacy (115, 116, 117), few studies have investigated the specific targets or pathways of m6A and ferroptosis in immune therapy. Given that ferroptosis and m6A play critical roles in tumors, further research on m6A-modified ferroptosis in diverse cancers is needed.
M6A and pyroptosis
Pyroptosis, a lytic form of cell death, is characterized by NLR pyrin domain containing 3 (NLRP3), apoptotic speck-like protein containing CARD (ASC), cleaved Caspase-1, Gasdermin-D (GsdmD) p30, IL-1β, and IL-18, which serve as important regulators of pyroptosis (120). Although an increasing number of studies have demonstrated the critical role of pyroptosis or m6A in different diseases, including different cancers (121), studies on the m6A-pyroptosis axis in cancer seem to be few, while most focus on ischemic diseases and some chronic diseases (Table 2). In hypoxic pulmonary hypertension, the degradation of lncRNA FENDRR mediated by YTHDC1 promotes HPAEC pyroptosis by regulating DRP1 promoter methylation (122). In slow-transit constipation, METTL3 promotes the pyroptosis of glutamic acid-induced ICCs by interacting with DGCR8 and modulating the miR-30b-5p/PIK3R2 axis in an m6A-dependent manner (123).
In terms of m6A-pyroptosis in ischemia-reperfusion injury, hypothermia protects neurons from cerebral ischemia-reperfusion injury by downregulating the secretion of the pyroptosis-related proteins NLRP3, ASC, and some pro-inflammatory factors by activating PI3K/Akt signaling via the m6A modification of PTEN mRNA (124). Meanwhile, METTL3 promotes pyroptosis in myocardial cells to exacerbate myocardial ischemia-reperfusion injury in an m6A-dependent manner (125). The m6A-pyroptosis axis seems to play an important role in ischemia-reperfusion injury, which is consistent with the results of a similar summary (126), but the regulation of m6A-pyroptosis at other sites of ischemia-reperfusion injury and more specific mechanisms remain to be explored.
M6A-ferroptosis axis in diverse cancers.
| Disease models | m6a regulation | Type of PCD | Biofunction | Reference |
|---|---|---|---|---|
| Glioblastoma | METTL3 | ferroptosis | RNA binding protein NKAP promotes SLC7A11 mRNA splicing in an m6A-dependent manner. | (109) |
| Thyroid Cancer | FTO | ferroptosis | FTO can inhibit the development of Thyroid cancer by downregulating SLC7A11 in m6A independently. | (110) |
| ALKBH5 | ferroptosis | ALKBH5 inhibits thyroid cancer progression by promoting ferroptosis through TIAM1-Nrf2/HO-1 axis. | (118) | |
| Non-Small Cell Lung Carcinoma | METTL3 | ferroptosis | miR-4443 in cisplatin-resistant NSCLC tumor tissue-derived exosomes regulated the expression of FSP1 in an m6A manner via METLL3. | (112) |
| Lung Adenocarcinoma. | YTHDC2 | ferroptosis | YTHDC2 suppressed SLC3A2 via inhibiting HOXA13 in an m6A-indirect manner. | (111) |
| Breast Cancer | METTL14 | ferroptosis | METTL14 regulated FGFR4 diminishes glutathione synthesis and Fe2+ efflux efficiency via the β-catenin/TCF4-SLC7A11/FPN1 axis. | (113) |
| Gastric Cancer | YTHDF2 | ferroptosis | lncRNA-CBSLR interacted with YTHDF2 to form a CBSLR/YTHDF2/CBS signaling axis to reduce the expression of ACSL4. | (114) |
| Hepatocellular Carcinoma | IGF2BP3 | ferroptosis | IGF2BP3-NRF2 axis regulates ferroptosis in hepatocellular carcinoma. | (119) |
| Hepatoblastoma | METTL3 | ferroptosis | METTL3-mediated SLC7A11 m6A modification enhances HB ferroptosis resistance in an IGF2BP1 dependent manner. | (108) |
Additionally, m6A-mediated pyroptosis occurs in some chronic diseases. In patients with diabetic nephropathy (DN), WTAP is highly expressed, and WTAP knockdown inhibits the m6A methylation of NLRP3 mRNA to downregulate NLRP3 inflammasome activation, which further induces cell pyroptosis and inflammation (127). Similarly, in diabetic nephropathy, the total flavones of Abelmoschus manihot (TFA) inhibit the pyroptosis of podocytes under high glucose conditions by regulating METTL3-dependent m6A modification and inhibiting the activation of the NLRP3 inflammasome and PTEN/PI3K/Akt signaling (128). Additionally, METTL3 expression is lower in T2DM patients. The overexpression of METTL3 alleviates high-glucose-induced apoptosis and pyroptosis in human retinal pigment epithelial (RPE) cells via the METTL3/miR-25-3p axis (129). The same mechanism is involved in diabetic cardiomyopathy. METTL3 degrades lncRNA TINCR mediated by YTHDF2, which further decreases the expression of NLRP3, a key pyroptosis-related protein, to regulate the occurrence of pyroptosis and diabetic cardiomyopathy (130). In diabetic retinopathy, circFAT1 interacts with YTHDF2, which increase the expression of LC3B to promote autophagy and inhibit pyroptosis in high glucose-induced retinal pigment epithelial (RPE) cells (131). The m6A-pyroptosis axis appears to play a profound role in the chronic complications of diabetes, but more research is needed to extend our understanding of the epigenetic regulation of pyroptosis in DCM progression. Interestingly, the m6A-pyroptosis is not restricted to epithelial cells and is involved in some immune cells. In atherosclerosis and acute coronary syndrome (ACS), IRF-1 represses circ-0029589 expression in an METTL3 dependent manner, thereby promoting macrophage pyroptosis and inflammatory responses (132). In addition, the METTL3/MALAT1/PTBP1/USP8/TAK1 axis in liver fibrosis promotes pyroptosis and macrophage M1 polarization, thereby exacerbating liver fibrosis progression (102). Although m6A-modified pyroptosis plays an important role in non-neoplastic diseases, its role in tumors requires further study.
M6A and necroptosis
Necroptosis is a form of programmed necrosis that occurs when apoptosis is blocked by extracellular signals (death receptor-ligand binding) or intracellular triggers (microbial nucleic acids) through a series of phosphorylation events that result in the production of pore complexes on the plasma membrane by MLKL, leading to the secretion of DAMP and subsequent cellular self-destruction (136). Necroptosis is characterized by organelle swelling, cell membrane rupture, and the disintegration of the cytoplasm and nucleus, with RIPK1, RIPK3, and MLKL as the main molecules involved (137). In addition, tumor necrosis factor (TNF), Toll-like receptor (TOLLR) family members, interferon, and other mediators have been demonstrated to act as key genes in necroptosis (138).
M6A-pyrpotosis axis in no-cancer diseases.
| Hypoxic pulmonary hypertension | YTHDC1 | pyroptosis | YTHDC1-induced decay of lncRNA FENDRR promotes HPAEC pyroptosis by regulating DRP1 promoter methylation. | (122) |
| Slow transit constipation | METTL3 | pyroptosis | METTL3 contributes to slow transit constipation by regulating miR-30b-5p/PIK3R2/Akt/mTOR signaling cascade through interacting with DGCR8. | (123) |
| Sepsis | YTHDF1 | pyroptosis | YTHDF1 alleviates sepsis by upregulating WWP1 to induce NLRP3 ubiquitination and inhibit caspase-1-dependent pyroptosis. | (133) |
| Liver fibrosis | METTL3 | pyroptosis | The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. | (134) |
| Atherosclerosis (AS) and acute coronary syndrome (ACS) | METTL3 | pyroptosis | IRF-1 can repress circ-0029589 expression in a METTL3-dependent manner, thereby promoting macrophage pyroptosis and inflammatory responses | (132) |
| Myocardial Ischemia-Reperfusion Injury | METTL3 | pyroptosis | METTL3 promotes DGCR8 binding to pri-miR-143-3p in an m6A dependent manner, thus enhancing miR-143-3p expression to inhibit PRKCE transcription and further aggravating cardiomyocyte pyroptosis and MI/R injury. | (125) |
| Cerebral ischemia/reperfusion (I/R) injury | m6A | pyroptosis | Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3β signaling pathway. | (124) |
| Diabetic nephropathy | WTAP | pyroptosis | WTAP promotes the expression of NLRP3 in a IGFBP2 dependent manner to upregulate NLRP3 inflammasome activation, which further induces cell pyroptosis and inflammation. | (127) |
| Diabetic nephropathy | METTL3 | pyroptosis | TFA can ameliorate pyroptosis by regulating the expression of METTL3 and regulating NLRP3-inflammasome activation and PTEN/PI3K/Akt signaling. | (128) |
| Diabetic cardiomyopathy (DCM) | METTL14 | pyroptosis | METTL14 suppresses pyroptosis and DCM via downregulating lncRNA TINCR, which further decreases the expression of key pyroptosis-related protein, NLRP3. | (135) |
| Diabetic retinopathy | YTHDF2 | pyroptosis | CircFAT1 interact with YTHDF2 to increase the expression of LC3B, thus promoting autophagy and inhibiting pyroptosis of RPE cells induced by HG. | (131) |
In CRC, patients resistant to oxaliplatin have higher METTL3 expression and infiltration of M2-type macrophages. Further studies revealed that the TRAF5-mediated inhibition of necroptosis contributes to METTL3-triggered OX resistance. This finding shows the role of the m6A-necroptosis axis in OX resistance and provides a new target for patients with OX resistance in CRC (139). In addition, necroptosis-related genes, such as necroptosis-related mRNA and necroptosis-related lncRNA, are indicators of a worse prognosis and correlate with m6A gene expression and immune function, which are used to predict the prognosis and immune response in different cancers; however, the specific mechanism remains need to be further investigated (140), (141), (142), (143). Few studies have examined the role of m6A modification in necroptosis; therefore, the precise regulatory mechanisms involved are still unknown.
Small molecular compounds targeting m6A modification in various cancers
The m6A modification has been found to act as a pivotal role in various cancers. Therefore, inhibitors and regulators targeting m6A regulators may be effective new approaches for cancer therapy. STM2457, a highly potent and selective first-in-class catalytic inhibitor of METTL3, was demonstrated to hinder the growth of AML without impacting normal hematopoiesis (144). It had also been proven to block the proliferation of intrahepatic cholangiocarcinoma in an m6A-YTHCF2-dependent manner (145). In addition to METTL3, other m6A regulators are also key targets for treating cancers with abnormal m6A levels. Rhein, competitively bounds to FTO or AlkB catalytic in vitro, displayed the enhancement of antiproliferative effects of atezolizumab based on breast cancer (4T1) regression (146). It also examined the inhibitory effect in breast cancer in vitro and in vivo (147). Meclofenamic acid (MA), a highly selective inhibitor of FTO, restored gefitinib sensitivity via FTO/m6A-Demethylation/c-Myc in Non-Small Cell Lung Cancer (148). It also played a protective effect in cisplatin-induced acute kidney injury (149). In glioma, MA2 (the ethyl ester form of meclofenamic acid) inhibited FTO and suppressed proliferation by increasing the effect of the chemotherapy drug temozolomide (150). MA2 could also rescue the cisplatin-induced cytotoxicity of bladder cancer cells (151). Subsequently, other FTO inhibitors, such as MO-I-500, FB23-2, R-2HG, CS1, and CS2, are also showed the antitumor effect on diverse cancers, including Alzheimer's disease (AD) (152), nasopharyngeal carcinoma (153), Breast Cancer (154), AML (155), (156), (157), cholangiocarcinoma (158), renal cell carcinoma (96). Furthermore, IGF2BP1 acts as the post-transcriptional super-enhancer of E2F-driven gene expression in cancer. The small molecule, BTYNB, could disrupt this enhancer function by impairing the IGF2BP1-RNA association. It also showed the inhibitory potency in the treatment of solid cancers (159). In conclusion, although the small molecular compounds targeting m6A modification for clinical application is still in the initial phase, it is expected that drugs targeting m6A modification will be improved and developed and finally be used for clinical treatment over the next few years with more understanding of the function and mechanism of m6A in cancer.
Potential therapeutic applications of m6A-modified PCD in cancers
Increasing evidence has gradually proven that an anti-tumor strategy based on PCD and m6A may solve some existing problems in anti-cancer therapies (Table 3). In chemotherapy efficacy and resistance, diverse types of regulators of PCD and m6A molecules show efficacy in the chemoresistance of tumor cells (160), (161). The combination of m6A and PCD also improves anti-cancer efficacy. In the m6A-apoptosis axis, WTAP knockdown facilitates cell apoptosis and inhibits cisplatin resistance in nasal-type natural killer/T-cell lymphoma (162). Conversely, FTO enhances chemoresistance in CRC through SIVA1-mediated apoptosis via a YTHDF2-dependent mechanism (163). In terms of m6A-autophagy, METTL3-mediated autophagy reverses gefitinib resistance in NSCLC cells by β-elemene (100). In seminomas, METTL3 regulates autophagy and sensitivity to cisplatin by targeting ATG5 (164). 5-Azacytidine, a methyltransferase inhibitor and anticancer drug, stimulates an autophagic response to sensitize cancer cells to drug responsiveness during hydrogen peroxide-induced oxidative stress in insulinoma β-TC-6 cells (165). Regarding the m6A-ferroptosis axis, METTL14-modified FGFR4 increases anti-HER2 resistance by inhibiting ferroptosis mediated by the β-catenin/TCF4-SLC7A11/FPN1 axis in recalcitrant HER2-positive breast cancer (113). The low level of hypoxia-inducible lncRNA-CBSLR manifests a worse clinical outcome and a poorer response to chemotherapy, and regulates ferroptosis and chemoresistance through m6A-YTHDF2-dependent modulation (114). In terms of the m6A-pyroptosis axis, recent research indicated that the lower expression levels of DFNA5/GSDME in most tumor cells than in normal cells is attributed to the methylation of mRNA, thus making it difficult to activate pyroptosis to increase the sensitivity of chemotherapeutic drugs in most tumor cells. Therefore, appropriate chemotherapeutic drugs can be selected based on the expression levels of DFNA5/GSDM, in order to increase their effects (166). In immune therapy, ALKBH5-dependent HMGB1 expression decreases hepatocyte apoptosis and mediates the STING-IFN regulatory factor 3 innate immune response in radiation-induced liver diseases (167). FTO promotes melanoma processing and anti-PD-1 resistance, and suggests the potential of the combination of FTO inhibition with anti-PD-1 blockade in resistance to immunotherapy (98).
In conclusion, m6A-PCD may play a critical role in drug efficacy and resistance, including chemotherapy resistance, immune efficacy, and drug side effects in m6A-apoptosis. An increasing number of studies on m6A-PCD have focused on m6A autophagy and m6A-apoptosis. However, few studies have investigated the therapeutic application of m6A-modified pyroptosis and necroptosis, and further studies are needed.
Summary
Emerging studies have shown that m6A modification affects apoptosis and autophagy, thereby influencing the development of diverse cancers. However, few studies have examined the effects of m6A modification on ferroptosis, pyroptosis, and necroptosis in cancer. M6A-pyroptosis mainly focuses on non-neoplastic diseases, while m6A-necroptosis has been rarely studied in cancer. In brief, these studies revealed that m6A plays a remarkably important role in PCD and tumor development. However, the influence of m6A modification on PCD remains largely unclear, similar to the contradictory role of m6A modification in diverse cancers. For instance, METTL3 acts as an apoptotic driver in LUAD (173) whereas WTAP serves as a suppressor in endometrial cancer (174). In addition, m6A modification serves as an autophagy driver in NSCLC (100) whereas METTL3 functions as a suppressor in HCC (175). Whether PCD regulated by m6A exerts a promotive or inhibitive effect may primarily depend on the level of m6A (the dynamic balance between writers and erasers), the different readers, the variety (mRNA or ncRNA), the function of target genes, and different diseases.
The potential clinical application of m6A-modified PCD
| M6A associated molecules | Target gene | Type of PCD | Disease models | Clinical application | Biofunction | Reference |
|---|---|---|---|---|---|---|
| IGF2BP1 | PDL1 | apoptosis | hepatocellular carcinoma | immunotherapy | The inhibition of IGF2BP1 inhibits the development of hepatocellular carcinoma through activating immune cells infiltration and blocking PD-L1 expression to regulate the tumor immune microenvironment. | (87) |
| METTL3 | COL10A1 | apoptosis | LUSC | immunotherapy | COL10A1 secreted by Cancer-associated fibroblasts (CAFs), upregulated by METTL3, can promote LUSC cell proliferation and repress apoptosis-induced oxidative stress. | (88) |
| ALKBH5 | HMGB1 | apoptosis | radiation-induced liver diseases (RILD) | immunotherapy | ALKBH5-modicated HMGB1 expression mediates STING-interferon regulatory factor 3 innate immune response. | (167) |
| FTO | PDK1 | apoptosis | glioblastoma multiforme | temozolomide chemoresistance | Long noncoding RNA just proximal to X-inactive specific transcript promotes stability of PDK1 mRNA in an m6A-dependent manner. | (168) |
| METTL14 | TRAF1 | apoptosis | renal cell carcinoma | sunitinib resistance | TRAF1 overexpression can promote sunitinib resistance by modulating apoptotic in a METTL14-dependent manner. | (89) |
| METTL3 | microRNA-221-3p | apoptosis | breast cancer | adriamycin resistance | METTL3 accelerates pri-microRNA-221-3p maturation in a m6A-dependent manner. | (169) |
| ALKBH5 | LncRNA CASC8 | apoptosis | esophageal squamous cells | cisplatin resistance | LncRNA CASC8 overexpression can activate the Bcl2/caspase3 pathway to decrease the cisplatin sensitivity of esophageal squamous cells. | (91) |
| METTL3 | TRIM11 | apoptosis | nasopharyngeal carcinoma | chemoresistance | METTL3-medicated TRIM11 promoted Daple ubiquitin-mediated degradation to upregulate β-catenin expression, thus inducing ABCC9 expression. | (170) |
| METTL3 | lncRNA SNHG17 | apoptosis | lung adenocarcinoma | gefitinib resistance | METTL3-induced lncRNA SNHG17 reduces the expression of LATS2. | (171) |
| METTL3 | miR-146a-5p | apoptosis | bladder cancer | Melittin resistance | METTL3-guided m6A modification can accelerate the pri-miR-146 maturation. | (172) |
| FTO | PD-1 | autophagy | melanoma | immunotherapy | FTO can increase the anti- PD-1 resistance of melanoma through the autophagy and NF-κB pathway. | (98) |
| METTL3 | β-elemene | autophagy | non-small cell lung cancer | gefitinib resistance | METTL3-mediated autophagy can reverse this gefitinib resistance by the regulation of β-elemene. | (100) |
| METTL3 | FOXO3 | autophagy | HCC | sorafenib resistance | METTL3 can improve the sorafenib resistance of HCC cells through stabilizing forkhead box class O3 (FOXO3) mediated by YTHDF1 and inhibiting the occurrence of autophagy. | (102) |
| METTL14 | FGFR4 | ferroptosis | breast cancer | anti-HER2 therapy | METTL14 medicated FGFR4 can reduce the resistance to anti-HER2 therapy through the activation of ferroptosis by blocking glutathione synthesis and Fe2+ efflux efficiency. | (113) |
| METLL3 | miR-4443 | ferroptosis | non-small cell lung cancer | cisplatin resistance | METLL3 medicated miR-4443 can regulate the expression of FSP1 to increase the resistance to cisplatin and promote tumor growth. | (112) |
More importantly, the mechanism of methylation modification of all m6A-related proteins is not the same, the diverse biological processes which be affected by m6A modification is not specificity, the target genes may be regulated by diverse readers, and the m6A-related proteins identified thus far may have more than one role beyond methylation, which may lead to diametrically opposed roles in disease.
Interestingly, in addition to m6A, which regulates PCD, it also affects the m6A levels. The release of neutrophil extracellular traps (NETs) activates ferroptosis depending on the METTL3-induced m6A modification of GPX4 in sepsis-associated acute lung injury (176). Meanwhile, NET-activated METTL3 leads to abnormal autophagy in sepsis-associated acute lung injury (177).
Moreover, except for the regulation of m6A in a single type of PCD, m6A indirectly regulates one type of PCD through another type of PCD. For example, in hepatic stellate cells, m6A-modified BECN1 promotes the activation of autophagy, thus inducing ferroptosis (178). M6A-medicated PCD may be an indirect way to influence the activation of other PCD signals. It is believed that further studies on PCD interactions will provide an explanation. Further studies are needed to explore the mutual link between m6A modification and PCD in different diseases.
In addition, m6A modification could also happen on TAM, especially in immune cells, to indirectly effect tumor development. The balance between cancer cells and TAM may account for the contradictory role of m6A and PCD. Studies have shown that RNA modification is involved in the development, differentiation, activation, migration, polarization and other biological processes of immune cells, thus regulating immune response and participating in the occurrence of some immune-related diseases (179). Such as m6A methyltransferase in TAMs promotes CD8+ T cell dysfunction and tumor progression (180). It also revealed that the immune cell can also occur PCD to influence the development of cancers. But the specific mechanisms that tie them together still require further study. Moreover, except for m6A, other chemical modifications in DNA, RNA, and protein are also irreplaceable, such as DNA methylation, m1A, m5C, ubiquitylation, Phosphorylation, lactation, glycosylation modification, and so on. Whether these epigenetic modifications play an indispensable role as m6A in PCD? Are the other types of PCD, such as immunogenic cell death and cuproptosis, also regulated by m6A or other epigenetic modifications? These questions remain needed to be further explored.
In general, m6A-associated targets would provide a new direction for clinical diagnosis, treatment, prognosis, and therapy resistance in cancer. In general, investigating the intricate relationship between m6A and PCD could improve our understanding of how certain diseases develop and lead to the development of new treatments.
Abbreviations
m6A: N6-methyladenosine methylation; PCD: programmed cell death; ncRNA: non-coding RNA; METTL3/14: methyltransferase-like protein 3/14; METTL5/16: methyltransferase-like protein 5/16; WTAP: Wilms tumor-associated protein; VIRMA: vir-like m6A methyltransferase associated; RBM15RNA: binding motif protein 15; MTC: methyltransferase complex; SAM: S-adenosylmethionine; ZC3H13: zinc finger CCCH domain-containing protein 13; FTO: obesity-associated protein; ALKBH3/5: α-ketoglutarate-dependent dioxygenase alkB family member 3/5; YTH: domain-containing family protein (YTHDF1-3 and YTHDC1-2); HNRNP: heterogeneous nuclear ribonucleoprotein; eIF3: eukaryotic translation initiation factor 3; IGF2BP1/2/3: insulin-like growth factor-2 mRNA-binding proteins 1/2/3; AML: acute myeloid leukemia; GBM: glioma; NPC: nasopharyngeal carcinoma; HCC: hepatocellular carcinoma; CRC: colorectal cancer; NSCLC: non-cellular lung cancer; ATGs: autophagy-related homolog; BC: breast cancer; Bcl-2: BCL2 apoptosis regulator; (KCNK15-AS1): potassium two pore domain channel subfamily K member 15 and WISP2 antisense RNA 1; GLUT1: glucose transporter 1; CAFs: cancer-associated fibroblasts; ATG: autophagy-related genes; ULK1: uncoordinated 51-like kinase 1; TFEB: transcription factor EB; pSS: Sjögren's syndrome; FOXO3: forkhead box class O3; NLRP3: NLR pyrin domain containing 3; ASC: apoptotic speck-like protein containing CARD; GsdmD: gasdermin-D; DN: diabetic nephropathy; TFA: total flavones of Abelmoschus manihot; RPE: retinal pigment epithelial; HG: high glucose; AS: atherosclerosis; ACS: acute coronary syndrome; NKTCL: nasal-type natural killer/T-cell lymphoma; 5-azaC: 5-azacytidine; RILD: radiation-induced liver diseases; NETs: neutrophil extracellular traps; GPX4: glutathione peroxidase 4; lncRNAs: long non-coding RNAs; LUAD: lung adenocarcinoma.
Author contributions
Q.T., M.Y. and J.C. organized the whole topic. J.C., M.Y., J.B. and C.H. drafted the manuscript. F.L., D.G. and P.Y. revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018Jan4;46(D1):D303-7
2. Han SH, Choe J. Diverse molecular functions of m6A mRNA modification in cancer. Exp Mol Med. 2020May;52(5):738-49
3. Qin S, Mao Y, Wang H, Duan Y, Zhao L. The interplay between m6A modification and non-coding RNA in cancer stemness modulation: mechanisms, signaling pathways, and clinical implications. Int J Biol Sci. 2021;17(11):2718-36
4. Imanishi M. Mechanisms and Strategies for Determining m6 A RNA Modification Sites by Natural and Engineered m6 A Effector Proteins. Chem Asian J. 2022Aug15;17(16):e202200367
5. Boulias K, Greer EL. Biological roles of adenine methylation in RNA. Nat Rev Genet. 2022 Oct 19
6. Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019Dec;18(1):103
7. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021Feb21;6(1):74
8. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P. et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018Mar;25(3):486-541
9. Zhao J, Jiang P, Guo S, Schrodi SJ, He D. Apoptosis, Autophagy, NETosis, Necroptosis, and Pyroptosis Mediated Programmed Cell Death as Targets for Innovative Therapy in Rheumatoid Arthritis. Front Immunol. 2021;12:809806
10. Duan HC, Wang Y, Jia G. Dynamic and reversible RNA N6 -methyladenosine methylation. Wiley Interdiscip Rev RNA. 2019Jan;10(1):e1507
11. Zhu W, Wang JZ, Wei JF, Lu C. Role of m6A methyltransferase component VIRMA in multiple human cancers (Review). Cancer Cell Int. 2021Mar17;21(1):172
12. Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021Jul21;49(13):7239-55
13. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016Jul21;63(2):306-17
14. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020Aug27;13(1):117
15. Shi B, Liu WW, Yang K, Jiang GM, Wang H. The role, mechanism, and application of RNA methyltransferase METTL14 in gastrointestinal cancer. Mol Cancer. 2022Aug16;21(1):163
16. Huang Q, Mo J, Liao Z, Chen X, Zhang B. The RNA m6A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis. 2022Oct7;13(10):852
17. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020May12;19(1):88
18. Turkalj EM, Vissers C. The emerging importance of METTL5-mediated ribosomal RNA methylation. Exp Mol Med. 2022Oct;54(10):1617-25
19. Ruszkowska A. METTL16, Methyltransferase-Like Protein 16: Current Insights into Structure and Function. Int J Mol Sci. 2021Feb22;22(4):2176
20. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018Mar1;32(5-6):415-29
21. Li Y, Su R, Deng X, Chen Y, Chen J. FTO in cancer: functions, molecular mechanisms, and therapeutic implications. Trends Cancer. 2022Jul;8(7):598-614
22. Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E. et al. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J Hematol Oncol. 2022Jan21;15(1):8
23. Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K. et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017Feb13;7:42271
24. Dai XY, Shi L, Li Z, Yang HY, Wei JF, Ding Q. Main N6-Methyladenosine Readers: YTH Family Proteins in Cancers. Front Oncol. 2021;11:635329
25. Shi R, Ying S, Li Y, Zhu L, Wang X, Jin H. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis. 2021Apr1;12(4):346
26. Liao S, Sun H, Xu C. YTH Domain: A Family of N6-methyladenosine (m6A) Readers. Genomics Proteomics Bioinformatics. 2018Apr;16(2):99-107
27. Xu Y, Zhang W, Shen F, Yang X, Liu H, Dai S. et al. YTH Domain Proteins: A Family of m6A Readers in Cancer Progression. Front Oncol. 2021;11:629560
28. Lu Y, Wang X, Gu Q, Wang J, Sui Y, Wu J. et al. Heterogeneous nuclear ribonucleoprotein A/B: an emerging group of cancer biomarkers and therapeutic targets. Cell Death Discov. 2022Jul25;8(1):337
29. Genuth NR, Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet. 2018Jul;19(7):431-52
30. Huang X, Zhang H, Guo X, Zhu Z, Cai H, Kong X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol. 2018Jun28;11(1):88
31. Wang J, Chen L, Qiang P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int. 2021Feb10;21(1):99
32. Lederer M, Bley N, Schleifer C, Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014Dec;29:3-12
33. Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D. et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. 2020Jan;22(1):54-66
34. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015Mar26;519(7544):482-5
35. Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J. et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther. 2020Jun3;28(6):1479-93
36. Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vågbø CB. et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021Jun10;184(12):3125-3142.e25
37. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017Nov2;45(19):11356-70
38. Li S, Qi Y, Yu J, Hao Y, He B, Zhang M. et al. Nuclear Aurora kinase A switches m6A reader YTHDC1 to enhance an oncogenic RNA splicing of tumor suppressor RBM4. Signal Transduct Target Ther. 2022Apr1;7(1):97
39. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014Jan2;505(7481):117-20
40. Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M. et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA. Mol Cancer. 2022Aug19;21(1):168
41. Chen M, Wei L, Law CT, Tsang FHC, Shen J, Cheng CLH. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018Jun;67(6):2254-70
42. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015Jun4;161(6):1388-99
43. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015Oct22;526(7574):591-4
44. Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J. et al. m6A Facilitates eIF4F-Independent mRNA Translation. Mol Cell. 2017Nov2;68(3):504-514.e7
45. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017May;27(5):626-41
46. Jin S, Li M, Chang H, Wang R, Zhang Z, Zhang J. et al. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKε/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol Cancer. 2022Apr9;21(1):97
47. Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L. et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020Jul2;27(1):64-80.e9
48. Fang R, Chen X, Zhang S, Shi H, Ye Y, Shi H. et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat Commun. 2021Jan8;12(1):177
49. Liu Z, He J, Han J, Yang J, Liao W, Chen N. m6A Regulators Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization In Nasopharyngeal Carcinoma. Front Immunol. 2021;12:762243
50. Liu H, Lyu H, Jiang G, Chen D, Ruan S, Liu S. et al. ALKBH5-Mediated m6A Demethylation of GLUT4 mRNA Promotes Glycolysis and Resistance to HER2-Targeted Therapy in Breast Cancer. Cancer Res. 2022Nov2;82(21):3974-86
51. Zhou B, Bie F, Zang R, Zhang M, Song P, Liu L. et al. RNA modification writer expression profiles predict clinical outcomes and guide neoadjuvant immunotherapy in non-small cell lung cancer. EBioMedicine. 2022Oct;84:104268
52. Song C, Zhou C. HOXA10 mediates epithelial-mesenchymal transition to promote gastric cancer metastasis partly via modulation of TGFB2/Smad/METTL3 signaling axis. J Exp Clin Cancer Res. 2021Feb9;40(1):62
53. Chen S, Yang C, Wang ZW, Hu JF, Pan JJ, Liao CY. et al. CLK1/SRSF5 pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and promotes growth and metastasis of pancreatic cancer. J Hematol Oncol. 2021Apr13;14(1):60
54. Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M. et al. JNK Signaling Promotes Bladder Cancer Immune Escape by Regulating METTL3-Mediated m6A Modification of PD-L1 mRNA. Cancer Res. 2022May3;82(9):1789-802
55. Liu L, Gu M, Ma J, Wang Y, Li M, Wang H. et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. 2022Jul20;21(1):149
56. Lin C, Ma M, Zhang Y, Li L, Long F, Xie C. et al. The N6-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol Cancer. 2022Mar19;21(1):80
57. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K. et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018Sep;20(9):1074-83
58. Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW. et al. WTAP-mediated m6A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022Jun;29(6):1137-51
59. Wang W, Shao F, Yang X, Wang J, Zhu R, Yang Y. et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N6-methyladenosine-dependent YTHDF binding. Nat Commun. 2021Jun21;12(1):3803
60. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X. et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2021Feb23;14(1):32
61. Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J. et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. 2021Jul17;14:112
62. Ma L, Xue X, Zhang X, Yu K, Xu X, Tian X. et al. The essential roles of m6A RNA modification to stimulate ENO1-dependent glycolysis and tumorigenesis in lung adenocarcinoma. J Exp Clin Cancer Res. 2022Jan25;41(1):36
63. Liu X, Gonzalez G, Dai X, Miao W, Yuan J, Huang M. et al. Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m6A-Based Epitranscriptomic Mechanism. Mol Ther. 2020Dec2;28(12):2593-604
64. Liu X, Yuan J, Zhang X, Li L, Dai X, Chen Q. et al. ATF3 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an N6-Methyladenosine-Based Epitranscriptomic Mechanism. Chem Res Toxicol. 2021Jul19;34(7):1814-21
65. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W. et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022Feb23;21(1):60
66. Zhang S, Zhao S, Qi Y, Li B, Wang H, Pan Z. et al. SPI1-induced downregulation of FTO promotes GBM progression by regulating pri-miR-10a processing in an m6A-dependent manner. Mol Ther Nucleic Acids. 2022Mar8;27:699-717
67. Xiao Q, Lei L, Ren J, Peng M, Jing Y, Jiang X. et al. Mutant NPM1-Regulated FTO-Mediated m6A Demethylation Promotes Leukemic Cell Survival via PDGFRB/ERK Signaling Axis. Front Oncol. 2022;12:817584
68. Qu J, Hou Y, Chen Q, Chen J, Li Y, Zhang E. et al. RNA demethylase ALKBH5 promotes tumorigenesis in multiple myeloma via TRAF1-mediated activation of NF-κB and MAPK signaling pathways. Oncogene. 2022Jan;41(3):400-13
69. Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN. et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021Jul10;14(1):109
70. Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong CC. et al. N6-Methyladenosine Reader YTHDF1 Promotes ARHGEF2 Translation and RhoA Signaling in Colorectal Cancer. Gastroenterology. 2022Apr;162(4):1183-96
71. Zhang N, Shen Y, Li H, Chen Y, Zhang P, Lou S. et al. The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp Mol Med. 2022Feb;54(2):194-205
72. Hu Y, Gong C, Li Z, Liu J, Chen Y, Huang Y. et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol Cancer. 2022Feb3;21(1):34
73. Fan HN, Chen ZY, Chen XY, Chen M, Yi YC, Zhu JS. et al. METTL14-mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022Feb14;21(1):51
74. He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C. et al. ALKBH5-mediated m6A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis. 2021Dec1;12(12):1121
75. Guo Z, Zhang X, Lin C, Huang Y, Zhong Y, Guo H. et al. METTL3-IGF2BP3-axis mediates the proliferation and migration of pancreatic cancer by regulating spermine synthase m6A modification. Front Oncol. 2022;12:962204
76. Yang X, Zhang S, He C, Xue P, Zhang L, He Z. et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020Feb28;19(1):46
77. Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M. et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022Jan10;41(1):19
78. Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J. et al. RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology. 2021Mar;160(4):1284-1300.e16
79. Krawiec K, Strzałka P, Czemerska M, Wiśnik A, Zawlik I, Wierzbowska A. et al. Targeting Apoptosis in AML: Where Do We Stand? Cancers (Basel). 2022Oct12;14(20):4995
80. Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014Jan;15(1):49-63
81. Van Opdenbosch N, Lamkanfi M. Caspases in Cell Death, Inflammation, and Disease. Immunity. 2019Jun18;50(6):1352-64
82. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017Nov;23(11):1369-76
83. Zhao F, Xu Y, Gao S, Qin L, Austria Q, Siedlak SL. et al. METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer's disease through aberrant cell cycle events. Mol Neurodegener. 2021Sep30;16(1):70
84. Du A, Li S, Zhou Y, Disoma C, Liao Y, Zhang Y. et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022May6;21(1):109
85. Yang Z, Cai Z, Yang C, Luo Z, Bao X. ALKBH5 regulates STAT3 activity to affect the proliferation and tumorigenicity of osteosarcoma via an m6A-YTHDF2-dependent manner. EBioMedicine. 2022Jun;80:104019
86. Yu T, Yao L, Yin H, Teng Y, Hong M, Wu Q. ALKBH5 Promotes Multiple Myeloma Tumorigenicity through inducing m6A-demethylation of SAV1 mRNA and Myeloma Stem Cell Phenotype. Int J Biol Sci. 2022;18(6):2235-48
87. Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK. et al. Allosteric Regulation of IGF2BP1 as a Novel Strategy for the Activation of Tumor Immune Microenvironment. ACS Cent Sci. 2022Aug24;8(8):1102-15
88. Li Y, Li X, Deng M, Ye C, Peng Y, Lu Y. Cancer-Associated Fibroblasts Hinder Lung Squamous Cell Carcinoma Oxidative Stress-Induced Apoptosis via METTL3 Mediated m6A Methylation of COL10A1. Oxid Med Cell Longev. 2022;2022:4320809
89. Chen Y, Lu Z, Qi C, Yu C, Li Y, Huan W. et al. N6-methyladenosine-modified TRAF1 promotes sunitinib resistance by regulating apoptosis and angiogenesis in a METTL14-dependent manner in renal cell carcinoma. Mol Cancer. 2022Dec;21(1):111
90. Wang Q, Chen C, Xu X, Shu C, Cao C, Wang Z. et al. APAF1-Binding Long Noncoding RNA Promotes Tumor Growth and Multidrug Resistance in Gastric Cancer by Blocking Apoptosome Assembly. Adv Sci (Weinh). 2022Oct;9(28):e2201889
91. Wu Q, Zhang H, Yang D, Min Q, Wang Y, Zhang W. et al. The m6A-induced lncRNA CASC8 promotes proliferation and chemoresistance via upregulation of hnRNPL in esophageal squamous cell carcinoma. Int J Biol Sci. 2022Jul18;18(13):4824-36
92. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008Dec;9(12):1004-10
93. Jin S, Zhang X, Miao Y, Liang P, Zhu K, She Y. et al. m6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018Sep;28(9):955-7
94. Chen X, Gong W, Shao X, Shi T, Zhang L, Dong J. et al. METTL3-mediated m6A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022Jan;81(1):87-99
95. Cui YH, Yang S, Wei J, Shea CR, Zhong W, Wang F. et al. Autophagy of the m6A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat Commun. 2021Apr12;12:2183
96. Xu Y, Zhou J, Li L, Yang W, Zhang Z, Zhang K. et al. FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int J Biol Sci. 2022;18(15):5943-62
97. Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H. et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021Feb23;6:76
98. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y. et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019Jun25;10:2782
99. Cheng L, Li H, Zhan H, Liu Y, Li X, Huang Y. et al. Alterations of m6A RNA methylation regulators contribute to autophagy and immune infiltration in primary Sjögren's syndrome. Front Immunol. 2022;13:949206
100. Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X. et al. The mechanism of m6A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020Nov11;11(11):969
101. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D. et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019May16;10(6):383
102. Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X. et al. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020Jun17;39(12):e103181
103. Stockwell BR. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022Jul7;185(14):2401-21
104. Dixon SJ. Ferroptosis: bug or feature? Immunol Rev. 2017May;277(1):150-7
105. Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021Sep;17(9):2054-81
106. Zhuang S, Ma Y, Zeng Y, Lu C, Yang F, Jiang N. et al. METTL14 promotes doxorubicin-induced cardiomyocyte ferroptosis by regulating the KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol Toxicol. 2021 Oct 14
107. Xu Y, Lv D, Yan C, Su H, Zhang X, Shi Y. et al. METTL3 promotes lung adenocarcinoma tumor growth and inhibits ferroptosis by stabilizing SLC7A11 m6A modification. Cancer Cell Int. 2022Jan7;22(1):11
108. Liu L, He J, Sun G, Huang N, Bian Z, Xu C. et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med. 2022May;12(5):e778
109. Sun S, Gao T, Pang B, Su X, Guo C, Zhang R. et al. RNA binding protein NKAP protects glioblastoma cells from ferroptosis by promoting SLC7A11 mRNA splicing in an m6A-dependent manner. Cell Death Dis. 2022Jan21;13(1):73
110. Ji FH, Fu XH, Li GQ, He Q, Qiu XG. FTO Prevents Thyroid Cancer Progression by SLC7A11 m6A Methylation in a Ferroptosis-Dependent Manner. Front Endocrinol (Lausanne). 2022;13:857765
111. Ma L, Zhang X, Yu K, Xu X, Chen T, Shi Y. et al. Targeting SLC3A2 subunit of system XC- is essential for m6A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med. 2021May20;168:25-43
112. Song Z, Jia G, Ma P, Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021Jul1;276:119399
113. Zou Y, Zheng S, Xie X, Ye F, Hu X, Tian Z. et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat Commun. 2022May13;13(1):2672
114. Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y. et al. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J Adv Res. 2022Mar;37:91-106
115. Xie H, Shi M, Liu Y, Cheng C, Song L, Ding Z. et al. Identification of m6A- and ferroptosis-related lncRNA signature for predicting immune efficacy in hepatocellular carcinoma. Front Immunol. 2022;13:914977
116. Zheng J, Guo J, Wang Y, Zheng Y, Zhang K, Tong J. Bioinformatic Analyses of the Ferroptosis-Related lncRNAs Signature for Ovarian Cancer. Front Mol Biosci. 2021;8:735871
117. Huang QR, Li JW, Yan P, Jiang Q, Guo FZ, Zhao YN. et al. Establishment and Validation of a Ferroptosis-Related lncRNA Signature for Prognosis Prediction in Lower-Grade Glioma. Front Neurol. 2022;13:861438
118. Li W, Huang G, Wei J, Cao H, Jiang G. ALKBH5 inhibits thyroid cancer progression by promoting ferroptosis through TIAM1-Nrf2/HO-1 axis. Mol Cell Biochem. 2022 Sep 7
119. Lu Z, Yang H, Shao Y, Sun W, Jiang Y, Li J. IGF2BP3-NRF2 axis regulates ferroptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2022Oct30;627:103-10
120. Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. 2022Jun20;7(1):196
121. Peng F, Liao M, Qin R, Zhu S, Peng C, Fu L. et al. Regulated cell death (RCD) in cancer: key pathways and targeted therapies. Signal Transduct Target Ther. 2022Aug13;7(1):286
122. Wang X, Li Q, He S, Bai J, Ma C, Zhang L. et al. LncRNA FENDRR with m6A RNA methylation regulates hypoxia-induced pulmonary artery endothelial cell pyroptosis by mediating DRP1 DNA methylation. Mol Med. 2022Oct25;28(1):126
123. Gong WJ, Li R, Dai QQ, Yu P. METTL3 contributes to slow transit constipation by regulating miR-30b-5p/PIK3R2/Akt/mTOR signaling cascade through DGCR8. J Gastroenterol Hepatol. 2022 Sep 6
124. Diao MY, Zhu Y, Yang J, Xi SS, Wen X, Gu Q. et al. Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3β signaling pathway. Brain Res Bull. 2020Jun;159:25-31
125. Wang X, Li Y, Li J, Li S, Wang F. Mechanism of METTL3-Mediated m6A Modification in Cardiomyocyte Pyroptosis and Myocardial Ischemia-Reperfusion Injury. Cardiovasc Drugs Ther. 2022 Jan 23
126. Wang J, Li Y, Zhang S. N(6)-methyladenosine modification: A vital role of programmed cell death in myocardial ischemia/reperfusion injury. Int J Cardiol. 2022Nov15;367:11-9
127. Lan J, Xu B, Shi X, Pan Q, Tao Q. WTAP-mediated N6-methyladenosine modification of NLRP3 mRNA in kidney injury of diabetic nephropathy. Cell Mol Biol Lett. 2022Jun27;27(1):51
128. Liu BH, Tu Y, Ni GX, Yan J, Yue L, Li ZL. et al. Total Flavones of Abelmoschus manihot Ameliorates Podocyte Pyroptosis and Injury in High Glucose Conditions by Targeting METTL3-Dependent m6A Modification-Mediated NLRP3-Inflammasome Activation and PTEN/PI3K/Akt Signaling. Front Pharmacol. 2021;12:667644
129. Zha X, Xi X, Fan X, Ma M, Zhang Y, Yang Y. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging (Albany NY). 2020May4;12(9):8137-50
130. Meng L, Lin H, Huang X, Weng J, Peng F, Wu S. METTL14 suppresses pyroptosis and diabetic cardiomyopathy by downregulating TINCR lncRNA. Cell Death Dis. 2022Jan10;13(1):38
131. Huang C, Qi P, Cui H, Lu Q, Gao X. CircFAT1 regulates retinal pigment epithelial cell pyroptosis and autophagy via mediating m6A reader protein YTHDF2 expression in diabetic retinopathy. Exp Eye Res. 2022Sep;222:109152
132. Guo M, Yan R, Ji Q, Yao H, Sun M, Duan L. et al. IFN regulatory Factor-1 induced macrophage pyroptosis by modulating m6A modification of circ_0029589 in patients with acute coronary syndrome. Int Immunopharmacol. 2020Sep;86:106800
133. Zhang S, Guan X, Liu W, Zhu Z, Jin H, Zhu Y. et al. YTHDF1 alleviates sepsis by upregulating WWP1 to induce NLRP3 ubiquitination and inhibit caspase-1-dependent pyroptosis. Cell Death Discov. 2022May4;8(1):244
134. Shu B, Zhou YX, Li H, Zhang RZ, He C, Yang X. The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. Cell Death Discov. 2021Nov27;7(1):368
135. Xie L, Yuan Y, Xu S, Lu S, Gu J, Wang Y. et al. Downregulation of hepatic ceruloplasmin ameliorates NAFLD via SCO1-AMPK-LKB1 complex. Cell Rep. 2022Oct18;41(3):111498
136. Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022Jan;8(1):21-7
137. Morgan MJ, Kim YS. Roles of RIPK3 in necroptosis, cell signaling, and disease. Exp Mol Med. 2022 Oct 12
138. Wei X, Xie F, Zhou X, Wu Y, Yan H, Liu T. et al. Role of pyroptosis in inflammation and cancer. Cell Mol Immunol. 2022Sep;19(9):971-92
139. Lan H, Liu Y, Liu J, Wang X, Guan Z, Du J. et al. Tumor-Associated Macrophages Promote Oxaliplatin Resistance via METTL3-Mediated m6A of TRAF5 and Necroptosis in Colorectal Cancer. Mol Pharm. 2021Mar1;18(3):1026-37
140. Wu Z, Huang X, Cai M, Huang P, Guan Z. Novel necroptosis-related gene signature for predicting the prognosis of pancreatic adenocarcinoma. Aging (Albany NY). 2022Jan24;14(2):869-91
141. Zou J, Lin Z, Jiao W, Chen J, Lin L, Zhang F. et al. A multi-omics-based investigation of the prognostic and immunological impact of necroptosis-related mRNA in patients with cervical squamous carcinoma and adenocarcinoma. Sci Rep. 2022Oct6;12(1):16773
142. Lin Z, Zou J, Sui X, Yao S, Lin L, Wang J. et al. Necroptosis-related lncRNA signature predicts prognosis and immune response for cervical squamous cell carcinoma and endocervical adenocarcinomas. Sci Rep. 2022Sep29;12(1):16285
143. Huang J, Xu Z, Teh BM, Zhou C, Yuan Z, Shi Y. et al. Construction of a necroptosis-related lncRNA signature to predict the prognosis and immune microenvironment of head and neck squamous cell carcinoma. J Clin Lab Anal. 2022Jun;36(6):e24480
144. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021May;593(7860):597-601
145. Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT. et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene. 2022Mar;41(11):1622-33
146. Shen Z, Zhu B, Li J, Qin L. Rhein Augments Antiproliferative Effects of Atezolizumab Based on Breast Cancer (4T1) Regression. Planta Med. 2019Oct;85(14-15):1143-9
147. Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L. et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019Mar28;18(1):46
148. Chen H, Jia B, Zhang Q, Zhang Y. Meclofenamic Acid Restores Gefinitib Sensitivity by Downregulating Breast Cancer Resistance Protein and Multidrug Resistance Protein 7 via FTO/m6A-Demethylation/c-Myc in Non-Small Cell Lung Cancer. Front Oncol. 2022;12:870636
149. Zhou P, Wu M, Ye C, Xu Q, Wang L. Meclofenamic acid promotes cisplatin-induced acute kidney injury by inhibiting fat mass and obesity-associated protein-mediated m6A abrogation in RNA. J Biol Chem. 2019Nov8;294(45):16908-17
150. Xiao L, Li X, Mu Z, Zhou J, Zhou P, Xie C. et al. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 2020Sep15;80(18):3945-58
151. Wen L, Pan X, Yu Y, Yang B. Down-regulation of FTO promotes proliferation and migration, and protects bladder cancer cells from cisplatin-induced cytotoxicity. BMC Urol. 2020Apr16;20(1):39
152. Cockova Z, Honc O, Telensky P, Olsen MJ, Novotny J. Streptozotocin-Induced Astrocyte Mitochondrial Dysfunction Is Ameliorated by FTO Inhibitor MO-I-500. ACS Chem Neurosci. 2021Oct20;12(20):3818-28
153. Huang WM, Li ZX, Wu YH, Shi ZL, Mi JL, Hu K. et al. m6A demethylase FTO renders radioresistance of nasopharyngeal carcinoma via promoting OTUB1-mediated anti-ferroptosis. Transl Oncol. 2023Jan;27:101576
154. Singh B, Kinne HE, Milligan RD, Washburn LJ, Olsen M, Lucci A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS One. 2016;11(7):e0159072
155. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H. et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019Apr15;35(4):677-691.e10
156. Qing Y, Dong L, Gao L, Li C, Li Y, Han L. et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. 2021Mar4;81(5):922-939.e9
157. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M. et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020Jul13;38(1):79-96.e11
158. Gao Y, Ouyang X, Zuo L, Xiao Y, Sun Y, Chang C. et al. R-2HG downregulates ERα to inhibit cholangiocarcinoma via the FTO/m6A-methylated ERα/miR16-5p/YAP1 signal pathway. Mol Ther Oncolytics. 2021Dec17;23:65-81
159. Müller S, Bley N, Busch B, Glaß M, Lederer M, Misiak C. et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020Sep4;48(15):8576-90
160. Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting cancer drug resistance: New insights from the perspective of N6-methyladenosine RNA modification. Drug Resist Updat. 2020Dec;53:100720
161. Lan Q, Liu PY, Bell JL, Wang JY, Hüttelmaier S, Zhang XD. et al. The Emerging Roles of RNA m6A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021Jul1;81(13):3431-40
162. Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF, Cheng KC. et al. Inflammation-related pyroptosis, a novel programmed cell death pathway, and its crosstalk with immune therapy in cancer treatment. Theranostics. 2021;11(18):8813-35
163. Lin Z, Wan AH, Sun L, Liang H, Niu Y, Deng Y. et al. N6-methyladenosine demethylase FTO enhances chemo-resistance in colorectal cancer through SIVA1-mediated apoptosis. Mol Ther. 2022 Oct 28;S1525-0016(22)00624-4
164. Chen H, Xiang Y, Yin Y, Peng J, Peng D, Li D. et al. The m6A methyltransferase METTL3 regulates autophagy and sensitivity to cisplatin by targeting ATG5 in seminoma. Transl Androl Urol. 2021Apr;10(4):1711-22
165. Filip K, Lewińska A, Adamczyk-Grochala J, Marino Gammazza A, Cappello F, Lauricella M. et al. 5-Azacytidine Inhibits the Activation of Senescence Program and Promotes Cytotoxic Autophagy during Trdmt1-Mediated Oxidative Stress Response in Insulinoma β-TC-6 Cells. Cells. 2022Apr4;11(7):1213
166. Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J. et al. The role of pyroptosis in cancer: pro-cancer or pro-"host"? Cell Death Dis. 2019Sep9;10(9):650
167. Chen G, Zhao Q, Yuan B, Wang B, Zhang Y, Li Z. et al. ALKBH5-Modified HMGB1-STING Activation Contributes to Radiation Induced Liver Disease via Innate Immune Response. Int J Radiat Oncol Biol Phys. 2021Oct1;111(2):491-501
168. Li XD, Wang MJ, Zheng JL, Wu YH, Wang X, Jiang XB. Long noncoding RNA just proximal to X-inactive specific transcript facilitates aerobic glycolysis and temozolomide chemoresistance by promoting stability of PDK1 mRNA in an m6A-dependent manner in glioblastoma multiforme cells. Cancer Sci. 2021Nov;112(11):4543-52
169. Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med. 2021Jan;53(1):91-102
170. Zhang R, Li SW, Liu L, Yang J, Huang G, Sang Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis. 2020May7;9(5):45
171. Zhang H, Wang SQ, Wang L, Lin H, Zhu JB, Chen R. et al. m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death Dis. 2022Jul28;13(7):657
172. Yan R, Dai W, Wu R, Huang H, Shu M. Therapeutic targeting m6A-guided miR-146a-5p signaling contributes to the melittin-induced selective suppression of bladder cancer. Cancer Lett. 2022May28;534:215615
173. Wu Y, Chang N, Zhang Y, Zhang X, Xu L, Che Y. et al. METTL3-mediated m6A mRNA modification of FBXW7 suppresses lung adenocarcinoma. J Exp Clin Cancer Res. 2021Mar6;40(1):90
174. Xie W, Liu N, Wang X, Wei L, Xie W, Sheng X. Wilms' Tumor 1-Associated Protein Contributes to Chemo-Resistance to Cisplatin Through the Wnt/β-Catenin Pathway in Endometrial Cancer. Front Oncol. 2021;11:598344
175. Li G, Deng L, Huang N, Cui Z, Wu Q, Ma J. et al. m6A mRNA Methylation Regulates LKB1 to Promote Autophagy of Hepatoblastoma Cells through Upregulated Phosphorylation of AMPK. Genes (Basel). 2021Oct30;12(11):1747
176. Zhang H, Liu J, Zhou Y, Qu M, Wang Y, Guo K. et al. Neutrophil extracellular traps mediate m6A modification and regulates sepsis-associated acute lung injury by activating ferroptosis in alveolar epithelial cells. Int J Biol Sci. 2022;18(8):3337-57
177. Qu M, Chen Z, Qiu Z, Nan K, Wang Y, Shi Y. et al. Neutrophil extracellular traps-triggered impaired autophagic flux via METTL3 underlies sepsis-associated acute lung injury. Cell Death Discov. 2022Aug27;8(1):375
178. Shen M, Li Y, Wang Y, Shao J, Zhang F, Yin G. et al. N6-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021Nov;47:102151
179. Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L. et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther. 2022Sep22;7:334
180. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J. et al. The loss of RNA N6-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8+ T cell dysfunction and tumor growth. Cancer Cell. 2021Jul12;39(7):945-957.e10
Author contact
![]() Corresponding author: Qiyun Tang, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO. 300 Guangzhou Road, Nanjing, China. Email: tqy831com
Corresponding author: Qiyun Tang, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO. 300 Guangzhou Road, Nanjing, China. Email: tqy831com

 Global reach, higher impact
Global reach, higher impact