10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(8):2304-2318. doi:10.7150/ijbs.83158 This issue Cite
Review
Methyltransferase Set7/9 as a Multifaceted Regulator of ROS Response
1. Institute of Cytology, Russian Academy of Sciences, 194064, St Petersburg, Russian Federation.
2. Institute of Fundamental Medicine and Biology, Kazan Federal University, 420008, Kazan, Russian Federation.
3. Institute of Pharmacology, University of Bern, 3010, Bern, Switzerland.
4. School of Medicine, Nazarbayev University, 010000, Astana, Kazakhstan.
Abstract
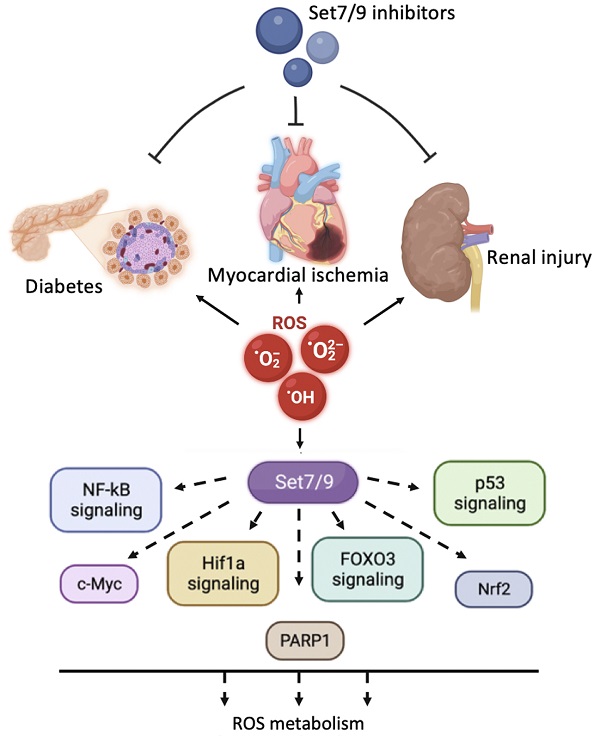
Reactive oxygen species (ROS) induce multiple signaling cascades in the cell and hence play an important role in the regulation of the cell's fate. ROS can cause irreversible damage to DNA and proteins resulting in cell death. Therefore, finely tuned regulatory mechanisms exist in evolutionarily diverse organisms that are aimed at the neutralization of ROS and its consequences with respect to cellular damage. The SET domain-containing lysine methyltransferase Set7/9 (KMT7, SETD7, SET7, SET9) post-translationally modifies several histones and non-histone proteins via monomethylation of the target lysines in a sequence-specific manner. In cellulo, the Set7/9-directed covalent modification of its substrates affects gene expression, cell cycle, energy metabolism, apoptosis, ROS, and DNA damage response. However, the in vivo role of Set7/9 remains enigmatic. In this review, we summarize the currently available information regarding the role of methyltransferase Set7/9 in the regulation of ROS-inducible molecular cascades in response to oxidative stress. We also highlight the in vivo importance of Set7/9 in ROS-related diseases.
Keywords: Set7/9, ROS metabolism, oxidative stress, p53, chromatin, diabetes, myocardial ischemic injury, nephropathy

 Global reach, higher impact
Global reach, higher impact