10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(10):3057-3076. doi:10.7150/ijbs.83368 This issue Cite
Review
Oncogenic SRSF3 in health and diseases
1. The State Key Laboratory Breeding Base of Basic Science of Stomatology & Key Laboratory of Oral Biomedicine Ministry of Education, School & Hospital of Stomatology, Wuhan University, Wuhan, Hubei, China.
2. Tumor Virus RNA Biology Section, HIV Dynamics and Replication Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland, USA.
Received 2023-2-9; Accepted 2023-5-30; Published 2023-6-12
Abstract
Serine/arginine rich splicing factor 3 (SRSF3) is an important multi-functional splicing factor, and has attracted increasing attentions in the past thirty years. The importance of SRSF3 is evidenced by its impressively conserved protein sequences in all animals and alternative exon 4 which represents an autoregulatory mechanism to maintain its proper cellular expression level. New functions of SRSF3 have been continuously discovered recently, especially its oncogenic function. SRSF3 plays essential roles in many cellular processes by regulating almost all aspects of RNA biogenesis and processing of many target genes, and thus, contributes to tumorigenesis when overexpressed or disregulated. This review updates and highlights the gene, mRNA, and protein structure of SRSF3, the regulatory mechanisms of SRSF3 expression, and the characteristics of SRSF3 targets and binding sequences that contribute to SRSF3's diverse molecular and cellular functions in tumorigenesis and human diseases.
Keywords: SRSF3, Alternative splicing, Cancer, Diseases
1. Introduction
The discovery of SRSF3 protein, also called SRp20 or SFRS3, goes back to a study of SR (serine/arginine rich) proteins with a mouse monoclonal anti-SR protein antibody, Mab104 in 1991 [1], which recognizes all phosphorylated SR proteins in calf thymus [2]. Subsequent sequence analysis showed that human SRSF3 gene is the same as mouse Srsf3 gene, which was previously called X16 [3]. SRSF3 is the smallest member of the splicing factor SR protein family which contains one or two N-terminal RNA recognition motifs (RRM) and a C-terminal serine and arginine-rich (RS) domain with consecutive RS repeats [4].
SRSF3, like other members of SR protein family, is considered as a key splicing factor in regulating constitutive and alternative pre-mRNA splicing and possibly RNA polyadenylation [5, 6]. SRSF3 associates with the active sites of RNA polymerase II (pol II) in the nucleus [7] and interacts with the C-terminal domain of pol II [8], thus, promoting coupling of transcription and RNA splicing. Along with application of Cre-loxP knockout technology, Jumaa et al. in 1999 found that SRSF3 is absolutely required for the development of embryos and the mice with Srsf3 knockout are often embryonic lethal [9]. Since then, more critical roles of SRSF3 have been revealed, including its roles in the development of human diseases and cancers. SRSF3 facilitates RNA export [10] by interacting with mRNA export receptor TAP/NXF1 [11] and is tumorigenic as a proto-oncogene when overexpressed [12]. SRSF3 promotes miRNA biogenesis by interacting and recruiting DROSHA to the Microprocessor cleavage site [13, 14]. Recent genome-wide approaches have identified numerous RNA targets of SRSF3 in a mouse tumor cell line [15] and a human osteosarcoma cell line [16]. This review will update what we have known about SRSF3 gene structure and expression, SRSF3 protein structure and functions, and most importantly its critical roles in tumorigenesis.
2. Characteristics of SRSF3 gene, mRNA, and protein
2.1 SRSF3 gene and mRNA
SRSF3 gene on human chr6p21.31 spans a 8422-bp region. Mouse Srsf3 gene on chr17 is 9423-bp long. Both contain seven exons and six introns, of which the exon 4 bearing a stop codon is an alternative exon (Figure 1). Both promoter transcription start sites and polyadenylation signals of human SRSF3 and mouse Srsf3 genes are now well defined [17]. We found that human SRSF3 promoter contains a TATA box at -26 bp and mouse Srsf3 promoter contains a TATA box at -27 bp upstream of each mapped transcription start site [17]. Further upstream of the TATA box are multiple consensus transcription factor binding sites including AP1, CREB, SP1, and E2F [18]. We discovered that both human SRSF3 gene and mouse Srsf3 gene have two polyadenylation signals (PAS), a major distal PAS “AUUAAA” and a minor proximal PAS “AAUAAA”. Because of exon 4 inclusion or exclusion and alternative usage of a distal or proximal PAS in pre-mRNA processing, human SRSF3 or mouse Srsf3 each expresses four different mRNA isoforms, of which the most abundant SRSF3 or Srsf3 mRNA is its isoform-1 mRNA less than ~1.5 kb in size [17].
The exon 4 in 456-nt size of both human SRSF3 and mouse Srsf3 is an alternative exon and contains a pre-mature stop codon [18, 19]. Thus, it is normally excluded from a mature SRSF3 mRNA during SRSF3 pre-mRNA splicing. Interestingly, the nucleotide sequence of exon 4 is highly conserved from human to mouse and chimpanzee, with 100% identity, but only 1-2 nt mismatches to most mammals, more than 97% to birds (Calypter anna et al.), and 91.92% to fish (Latimeria chalumnae) (Figure S1), suggesting that the exon 4 in all animals is excluded during SRSF3 pre-mRNA splicing. The SRSF3 long isoform with exon 4 inclusion is mainly degraded by non-sense mediated decay (NMD) [20, 21] and thus, only a fraction encodes a truncated SRSF3 protein if survived from NMD. In contrast, the exon 4-skipped SRSF3 mRNA is the major isoform of the RNA and has an open reading frame of 495 nt to encode a full-length SRSF3 protein (Figure 1). SRSF3 protein is well conserved among different animal species during evolution.
2.2 SRSF3 protein
2.2.1 The structure of SRSF3 protein
Full-length SRSF3 protein contains 164 amino acid (aa) residues and bears a N-terminal RNA recognition motif (RRM) and a C-terminal arginine/serine-rich (RS) domain (Figure 1). Like other SR protein family members, the RRM domain of SRSF3 provides RNA-binding specificity, while the RS domain is involved in protein-protein interactions. The RRM (1-86 aa residues) of SRSF3 (SRp20) has a βαββαβ topology in a structure very similar to SRSF7 (9G8). Solution structure of the SRSF3 RRM and of the RRM in complex with the RNA sequence CAUC showed all 4 nts in contacting with SRSF3 RRM in a semi-sequence-specific manner with only the 5' cytosine recognized specifically by the SRSF3 RRM [22]. A short arginine-rich peptide (aa 84-90, EKRSRNR) adjacent to the SRSF3 RRM, which does not contact RNA, is essential for interaction with an export factor Tip-associated protein TAP (NXF1) [22] to mediate RNA export [11].
Gene, mRNA, and protein structures of human SRSF3. Human SRSF3 gene contains seven exons and six introns, with exon 7 bearing two alternative poly(A) signals. Exon 4 is an alternative exon with a pre-mature stop codon. Because of alternative splicing and polyadenylation, SRSF3 gene expresses four mRNA isoforms. Full-length SRSF3 protein (164 amino acid residues) is encoded by transcripts without exon 4, whereas the transcripts containing the exon 4 encode a truncated SRSF3 protein (124 amino acid residues).
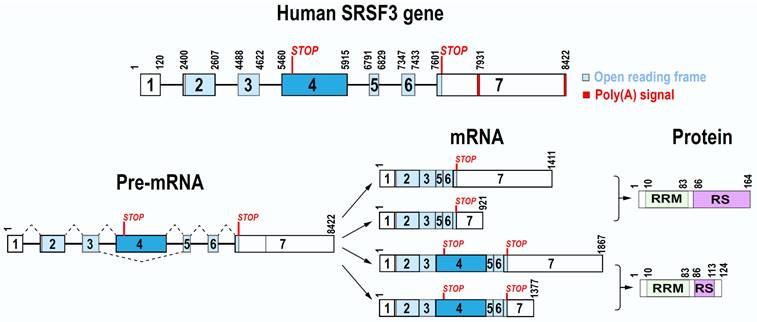
As stated above, the SRSF3 exon 4 contains a stop codon, inclusion of this exon 4 in SRSF3 mRNA leads to encode a truncated, non-functional SRSF3 protein containing only a small part of the RS domain [18]. Serum starving or oxidative stress induces exon 4 inclusion during SRSF3 pre-mRNA splicing (See description below) [18, 23].
2.2.2 Conservation of SRSF3 protein
SRSF3 protein is well conserved almost in all species of animal kingdom (Figure S2). By analyzing the conservation of SRSF3 protein sequences, impressively, we found that SRSF3 protein sequences are exactly the same in most mammals, including human, chimpanzee, monkey, dog, pig, fox, sheep, horse, cat, mouse, rat, beaver, and blue whale, except the hamster which has one aa substitution in the position of aa 28. Moreover, the birds, including sparrow, owl, chicken, and swan, share exactly the same SRSF3 protein sequences as human. As to reptilia, turtle also has the same SRSF3 protein sequence as human, while huge komodo dragon and small python all have only one substitution in aa 162, which is almost at the end of SRSF3 protein. Surprisingly, amphibian, exampled by caecilian or toad here, also have almost the same SRSF3 protein sequence as human with only a few aa substitutions, of which are mainly localized at the C-terminus.
2.2.3 SRSF3 protein turnover and stability
SRSF3 protein stability is regulated by ubiquitination or neddylation. SRSF3 has at least two ubiquitinated sites, Lys23 and Lys85 [24] and the ubiquitinated SRSF3 can be recognized by a monoclonal antibody against di-glycine-containing isopeptides. Under stress condition, SRSF3 can be neddylated at Lys11 and degraded by proteasome [25]. However, the neddylation in SRSF3 Lys85, which is also responsible for stress granule assembly [26], did not affect SRSF3 protein stability [25].
2.2.4 Phosphorylation of SRSF3 protein
In its steady state, SRSF3 protein is hypophosphorylated. Like other SR proteins, only under hypophosphorylated state, SRSF3 can bind NXF1 and promote RNA export [27, 28]. However, phosphorylation status of SRSF3 is important for its functions and cellular localization. Phosphorylation is required for SRSF3 nuclear import and participating RNA splicing. SRSF3 protein are phosphorylated at around 11-12 serine residues in its RS domain by serine-arginine protein kinase 2 (SRPK2). The SRSF3 RS domain also contains the Akt consensus site (RxRxxS/T) [29] and thus can be phosphorylated by the PI3K-Akt signal pathway [30]. Phosphatase 1 catalytic subunit α (PP1α) dephosphorylates SRSF3 [31]. SRSF1 (SF2/ASF) has two RRM domains, RRM1 and RRM2. Phosphoryl content of the SRSF1 RS domain could be protected by the RRM2 domain from dephosphorylation by phosphatases. SRSF3 lacks the extra RRM domain, and its phosphoryl content is more sensitive to phosphatases than SRSF1, suggesting a relative rapid dynamic switch of its phosphorylation status in correlation with the dynamic nature of spliceosome [31]. SRSF3 protein can be also dephosphorylated by PPM1G (also named PP2Cγ), a serine/threonine protein phosphatase, in hepatocellular carcinoma cells [32], although PPM1G-mediated dephosphorylation is involved in formation of the spliceosome [33].
2.2.5 Shuttle of SRSF3 between the nucleus and cytoplasm
SRSF3 protein mainly stays in the nucleus revealed by immunohistochemistry, immunocytochemistry, and nuclear and cytoplasmic fractionation analyses (exampled by ref. [34, 35]). However, similar to SRSF1 and SRSF7, SRSF3 does shuttle between the nucleus and cytoplasm, which facilitates mRNA export and stability in the cytoplasm. The phosphorylated RS domain and stable RNA binding mediated by the RRM domain of SRSF3 are required for shuttling [36]. Phosphorylated RS domain is recognized and imported into the nucleus by transportin-SR2 (TRN-SR2) [37]. SRSF3 RS domain contains not only a nuclear localization signal, but also directs itself to nuclear speckles [38].
Under certain conditions, SRSF3 may distribute to the cytoplasm. Viruses can promote cytoplasmic localization of SRSF3 to meet their requirements. For example, poliovirus 2A proteinase and human rhinovirus 16 2A proteinase can induce cytoplasmic relocalization of SRSF3 possibly by their cleavage of nuclear pore complex proteins [39]. Under osmotic stress condition, SRSF3 interacts with activated p38α MAPK in cardiomyocytes, resulting in partial SRSF3 distribution in the cytoplasm [40].
2.2.6 SRSF3 binds to chromatin
When SRSF3 stays in the nucleus, SRSF3 mainly participates RNA splicing in active sites of RNA polymerase II transcription [7]. In addition, it also binds to chromatin and both SRSF3 and SRSF1 are associated with interphase chromatin through histone H3. When histone H3 serine 10 is phosphorylated during mitosis, SRSF3 and SRSF1 are disassociated with hyperphosphorylated mitotic chromosomes, but re-associated with chromatin in the late M-phase. Phosphorylation of the RS domain of SRSF3 is required for its association with chromatin [41]. We found that SRSF3 knockdown induced cell cycle G2/M arrest [12], which may be partially related to its binding to chromatin. However, the exact roles of SRSF3 in association with chromatin remains to be determined.
DNA methylation affects the splicing of ~20% genomic alternative exons [42]. High DNA methylation in the gene body regions are associated with increased inclusion of alternatively spliced exons [43]. Histone H3 plays central roles in the association of chromatin with spliceosome as well SRSF3. Heterochromatin protein 1 (HP1) binds H3K9me3 in the chromatin and recruit SRSF3, which is transferred to the pre-mRNA and regulates alternative splicing [42]. H3K9 demethylase KDM3A is phosphorylated by protein kinase A (PKA) and recognizes H3K9 methylation to recruit SRSF3 to its target pre-mRNA, however, independent of its demethylase activity [44].
2.3 Homology between SRSF3 and other SR family members
So far, at least 12 SR proteins, SRSF1 to SRSF12 (SRrp35), have been discovered. All SR proteins have one or two N-terminal RRM domains and one C-terminal RS domain. Even though it is the smallest SR protein, SRSF3 can functionally complement splicing-deficient cytoplasmic S100 extracts [5, 45]. SRSF3 RS domain contains 14 “RS” or “SR” dipeptide repeats, which is similar to other members. However, the RRM domain of SRSF3 is significantly different from most of other members, which may suggest the RRM specificity for SRSF3. A few residues are conserved across SR family, such as Phe30, Gly34, Val51, Phe53, Asp55, Ala59, and Ala62. The sequence of “FAFVEFEDPRDAADA” in the middle of RRM domain is conserved in most members, except SRSF11 (p54). Interestingly, the RRM domain of SRSF3 is around 75% identity to that of SRSF7, suggesting that SRSF3 may recognize similar RNA motifs as SRSF7 (Figure S3).
3. SRSF3 expression and regulatory mechanisms
According to the data of human protein atlas (HPA) project, SRSF3 transcripts could be detected in almost all 27 different normal tissues, even though the expression levels are quite low in pancreas and salivary gland tissues [46] as showing in the webpage of human SRSF3 gene in Genbank. In mouse, the expression levels of SRSF3 is also significantly different in various tissues. Thymus, testis and spleen have high SRSF3 expression, while heart, liver and kidney have low or undetectable SRSF3 [3].
SRSF3 expression is absolutely required for embryonic development. Knockout of Srsf3 in mice blocked embryonic development before the blastocyst stage [9]. Conditional Srsf3 knockout in cardiomyocytes is lethel during embryonic development [47] and in neural crest cells affects mouse facial processes [30] SRSF3 expression has been reported in many types of cells, and is essential for cell proliferation [12, 48] However, overexpression of SRSF3 is tumorigenesis [12] Therefore, SRSF3 expression is regulated sophisticatedly and has to be maintained in balance in different cell types and at different stage of cell cycles (Figure 2).
3.1 SRSF3 promoter and its transcription regulation
Although SRSF3 promoter and its enhancers/silencers have not been well studied, several transcription factors or co-factors have been demonstrated to enhance SRSF3 gene transcription through its promoter. β-catenin is often mutated and escapes from degradation in tumors. Increased β-catenin translocates into the nucleus and interacts with TCF/LEF transcription factor family members to promote target gene expression and tumorigenesis [49]. A constitutively active mutant β-catenin can significantly increase SRSF3 expression by activating its transcription through the binding of TCF4 to an undefined region between nucleotides -225 and -1282 of SRSF3 promoter [50]. β-catenin is a central mediator of Wnt signaling pathway and CD133+ (a stemness marker for cancer stem cells) colon cancer stem cells express more SRSF3 than CD133- non-stem cancer cells. Activation of Wnt signaling pathway by Wnt3a protein significantly increases SRSF3 expression in colon cancer cells [51].
Other signal pathways related to transcriptional regulation have been explored. TGF-β1 enhances phosphorylation of p38 MAPK, which recruits NF-κB p65 binding to the SRSF3 promoter to increase SRSF3 expression [52]. Hepatocyte growth factor induces AKT phosphorylation to downregulate SRSF3 [53]. Insulin induces the levels of all phosphorylated SR proteins, including SRSF3, in rat hepatocytes [54], but promotes SRSF3 protein degradation by proteasome in human myeloblastic HL-60 cells [55]. Consistently, insulin-like growth factor 2 (IGF2) activates AKT (pSer473) and ERK1/2 (pThr202/Tyr204) phosphorylation to reduce SRSF3 expression in HepG2 cells. The IGF2 effect was specific for SRSF3 as other SR proteins did not change in expression [56]. Phospholipase C-β1 (PLC-β1) is a key nucleic enzyme for cell proliferation and differentiation by promotion of G1 cell cycle progression [57]. PLC-β1 interacts with SRSF3 directly in the nucleus to decrease SRSF3 protein level in Friend erythroleukemia cells [58], whereas hypoxia induces SRSF3 expression in human head and neck squamous cells [59]. HPV16 infection increases SRSF3 expression in undifferentiated keratinocytes through E2 protein [60] binding to SRSF3 promoter [61], but the increased SRSF3 promotes RNA splicing of viral early transcripts to disrupt integrity of the E2 ORF and therefore repress E2 expression, providing a negative feedback control of SRSF3 expression [62].
Regulatory mechanisms of SRSF3 expression. Expression of SRSF3 is regulated at the transcription level by transcription factors or at the post-transcriptional levels by alternative splicing of exon 4.
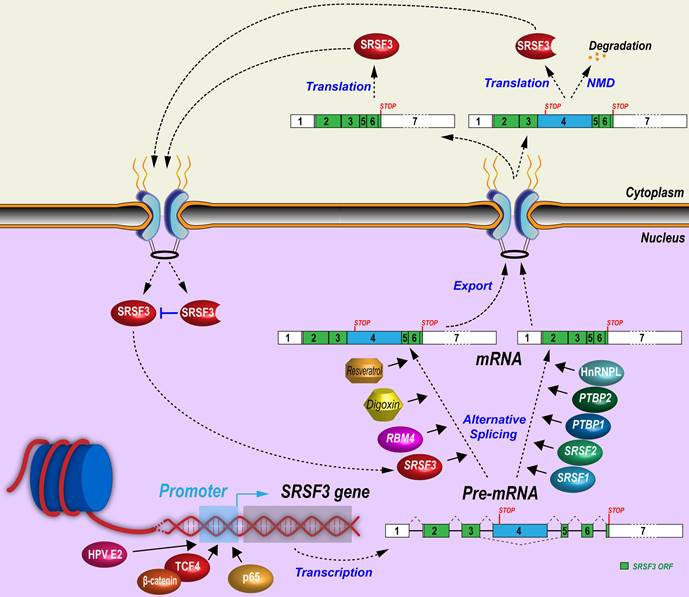
3.2 Alternative splicing of poison exon 4
Alternative splicing of the exon 4 bearing a stop codon is a very efficient way in regulating SRSF3 expression. Similar to SRSF3 alternative exon 4, most of other SR family members contain an alternative “poison” exon to negatively regulate their expression [19]. SRSF3 mRNAs with exon 4 inclusion encode a truncated SRSF3 protein. Only SRSF3 mRNAs with exon 4 exclusion can encode a full-length SRSF3 protein. Therefore, the increasing inclusion of exon 4 in SRSF3 RNA splicing will reduce the expression of full-length SRSF3 protein.
A number of mechanisms controlling SRSF3 exon 4 splicing have been revealed. First, SRSF3 itself can promote exon 4 inclusion and form a negative feedback regulation. Overexpression of Srsf3 significantly increases its own exon 4 inclusion in K46 cells (a murine B-cell lymphoma) [63]. Second, many other splicing factors regulate exon 4 splicing. In K46 cells, oncogenic SRSF1 and SRSF2 (SC35) suppress exon 4 inclusion and lead to the increase of SRSF3 protein [63, 64], but in U2OS cells, SRSF1 promotes inclusion of SRSF3 exon 4 [16]. A circRNA derived from SMARCA5 gene, cicrSMARCA5 which is not characterized carefully with unknown copy number and cellular locations, appears to increase the inclusion of SRSF3 exon 4 in glioblastoma cells [65]. Oncogenic splicing factors PTBP1 and PTBP2 suppress exon 4 inclusion via an exonic splicing suppressor, “UUUCU” [21]. In contrast, tumor suppressor RBM4 [66] reverses PTBP1-mediated skipping of exon 4 inclusion via an intronic CU-element [67]. In hepatocytes and in PLC/PRF/5 and H358 cells, splicing factor SLU7 inhibits inclusion of SRSF3 exon 4 [68, 69]. HnRNP L promotes SRSF3 full-length protein expression also by inhibiting exon 4 inclusion in oral squamous cell carcinoma cells [70]. Third, exon 4 splicing is associated with cell proliferation and stress. Mouse Srsf3 isoform with a poison exon 4 inclusion was high in starved K46 cells, and quickly decreased after serum stimulation [18]. Oxidative stress-induced by arsenite inhibits NMD to promote stability of SRSF3 isoform RNA with exon 4 inclusion in a human colon cancer cell line, HCT16 cells [23].
Other chemicals/drugs regulating exon 4 splicing have been reported. Digoxin is a cardiac glycoside drug used in the treatment of heart failure and arrhythmia. Digoxin represses SRSF3 expression by promoting SRSF3 exon 4 inclusion in a dose- and time-dependent manner [71]. Resveratrol is a small biocidal molecule widely produced, upon stress, by plants, including grape and peanut. Resveratrol and its derivatives show promising therapeutic effects in cancer, cardiovascular diseases, neurodegenerative diseases, infections, and so on [72]. Interestingly, resveratrol treatment of HEK293 cells increased SRSF3 exon 4 inclusion in a dose-dependent manner [73].
4. Function and molecular targets of SRSF3
4.1 Alternative RNA splicing targets of SRSF3 (Table 1)
SRSF3 interacts with RNA motifs to regulate RNA alternative splicing. SRSF3 also interacts with the C-terminal domain (CTD) of RNA polymerase II to regulate transcription-coupled alternative RNA splicing. Both SRSF1 and SRSF3 regulate the inclusion of an alternative exon of fibronectin gene, but deletion of the Pol II CTD was found only affecting the regulatory function of SRSF3, not SRSF1 [8].
Targets and/or binding motifs of SRSF3
| Gene or viral RNA | Exon or intron | Binding motif sequence | Function of SRSF3 | Pathways | Ref. |
|---|---|---|---|---|---|
| BCL-2 interacting cell death suppressor (BIS) | Exon 3 | UCAUCCUCAUC | Enhancing exon inclusion | Apoptosis | [178] |
| BPV-1 | Late RNA exon 2 | CACCACCACC | Enhancing exon 2 inclusion | BPV-1 L1 | [34] |
| Calcitonin/calcitonin gene-related peptide | Intron 4 | CUCCGCUCCUCUUCCAGGUAAGAC | Enhancing exon inclusion | CGRP (Calcitonin gene-related peptide) | [6] |
| CASP2 | Exon 8 | UCUUCAUC | Enhancing exon skipping | Apoptosis | [126] |
| CD44 | Exon v9 | CUUCGAUCAACGCCACGCCA | Enhancing exon inclusion | CD44 | [45] |
| CHK1 | Exon 3 | / | Enhancing exon inclusion | Cell cycle control | [16] |
| CKLF | Exon 3 | / | Enhancing exon inclusion | Inflammation | [16] |
| CLINT1 | Exon 11 | / | Enhancing exon inclusion | Vesicle-mediated transport | [16] |
| CPEB2 | Exon 4 | CAUCC | Enhancing exon inclusion | Epithelial to mesenchymal transition (EMT) | [113] |
| DDX5 | Exon 12 | / | Enhancing exon inclusion | β-catenin | [16] |
| EP300 | Exon 14 | CCAGCC | Enhancing exon inclusion | Chromatin remodeling | [16] |
| FOXM1 | Exon 9 | / | Enhancing exon skipping | Cell cycle progression | [12] |
| G6PD | Exon 12 | AGCCCCAGCC | Enhancing exon inclusion | Insulin | [54] |
| HER2 | Exon 15 | UGAUG, AGATG | Promoting upstream or downstream exon skipping) | HER2 signaling | [84] |
| HPV16 early RNA | Exon 2 | CACCGGAAACCCCUGCCACACCAC | Enhancing exon 2 inclusion | HPV16 E6/E7 | [34] |
| HPV18 early RNA | Exon 2 | ACCGCA, CCAGAC | Enhancing exon 2 inclusion | HPV18 E6/E7 | [62] |
| H2al1k | Exon 1 | ACAACAAGAAGACGCGCAUCAU | RNA export | Chromatin assembly | [10] |
| ILF3 | Exon 18 | AUCAACUUUUUACUCCAAUUUCCUCCA and UUAACUUCUCCCGCCUCUUGUAAUG | Enhancing exon 18 inclusion | Serine-Glycine-One-Carbon (SGOC) pathway | [80] |
| INSR | Exon 11 | CUCUUC | Enhancing exon inclusion | Insulin | [92] |
| KIF23 | Exon 18 | / | Enhancing exon inclusion | β-catenin | [16] |
| MAP4 | Exon 10 | / | Enhancing exon inclusion | Cell cycle progression | [16] |
| MAP4K4 | Exon 16 | GUCUUUUUCCA | Enhancing exon inclusion | RBM4/SRSF3/MAP4K4 | [67] |
| MBNL1 | Exons 5 and 7 | CAACC, CACCCGC | Enhancing exon inclusion | Expanded dsCUG binding and RNA splicing | [94] |
| MELK | Exon 11 | / | Enhancing exon inclusion | Cell cycle progression | [16] |
| NANOG | Exon 1 | CCCUUC, GCAUCG, CAUC | Nuclear export | Reprogram | [103] |
| PABPC1 | Exon 10 and 11 | / | Enhancing exon inclusion | mRNA metabolism | [16] |
| PKM | Exon 10 | AUCGUCC | Enhancing exon inclusion | Energy metabolism | [135] |
| PKP4 | Exon 7 | / | Enhancing exon inclusion | Rho GTPases pathway | [16] |
| PUS3 | Exon 3 | / | Enhancing exon inclusion | tRNA processing | [16] |
| SMC2 | Exon 3 and 4 | / | Enhancing exon inclusion | Cell cycle progression | [16] |
| SRSF1 | Exon 4 | / | Enhancing exon inclusion | Splicing factor | [16] |
| SYNGAP1 | Exon 14 | CCUCAAC | Enhancing exon skipping | Ras-Raf-MEK-ERK pathway | [95] |
| TP53 | Novel exon | UUUCAAA, UACUUGAC, and UACUUCCU | Enhancing exon skipping | TP53 | [48] |
| In vitro screening | CUCKUCY | Exonic splicing enhancer | [88] | ||
| In vivo iCLIP | UCAUC, CUUCA, CAUCA, CAUCU, and UCAAC, | Binding motifs | [87] |
/, unknown; K, guanosine or uridine; Y, cytidine or uridine.
4.1.1 High throughput analysis of SRSF3 targets and binding motifs
As an important splicing factor, SRSF3 regulates the alternative RNA splicing of hundreds of target genes. High-throughput methods were used to identify SRSF3 targets and binding motifs. We performed a genome-wide analysis of SRSF3 target genes and alternative RNA splicing events by using a human splicearray platform in human osteosarcoma U2OS cells with versus without SRSF3 knockdown [16]. We showed that SRSF3 regulates the transcription of 60 genes, including ERRFI1 [74], ANXA1 [75] and TGFB2 [76], and 182 RNA splicing events in 164 genes, including EP300, PUS3, CLINT1, PKP4, KIF23, CHK1, SMC2, CKLF, MAP4, MBNL1, MELK, DDX5, PABPC1, MAP4K4, SP1 and SRSF1 [16]. Many SRSF3 target genes and their alternative RNA splicing events are significantly associated with tumorigenesis, such as EP300 [77], mitogen-activated protein 4 kinase 4 (MAP4K4) [78], and SRSF1 [79]. Motif analysis showed that two SRSF3-binding motifs, CCAGC(G)C and A(G)CAGCA, are enriched to the RNA alternative exons. For example, SRSF3 promotes EP300 exon 14 inclusion through motif CCAGCC [16]. Later, we performed another genome-wide analysis of SRSF3 target genes and alternative splicing events by using a mouse splicearray platform in mouse fibroblast MEF3T3 cells with SRSF3 overexpression. The top lists also showed that SRSF3 regulated the expression of 11 genes and 139 splicing events in 134 genes [80]. Surprisingly, among these targets, alternative RNA splicing events in expression of ILF3 (interleukin enhancer binding factor 3) gene are the only conserved target splicing events of SRSF3 from human U2OS cells to mouse MEF3T3 cells.
RNA-seq has been widely used to analyze the effects of Srsf3 depletion on alternative splicing of mouse neonatal cardiomyocytes [47], neural crest cells [30], and mouse oocytes [81], and the roles of SRSF3 in development of human glioblastoma [82, 83]. Conditional knockout of Srsf3 in mouse oocytes led to sterility. Single cell RNA-seq found a number of unannotated splicing events in mouse Srsf3 knockout oocytes, the depression of some transposable elements, and some known alternative splicing events of the genes related to oocytes maturation, such as exon 11 skipping of Brd8 and mutually exclusive of Pdlim7 exon 5 and 6 [81]. High level of SRSF3 expression in glioblastoma has been noticed [80] and is essential for survival of glioblastoma cells [82, 83]. SRSF3 in glioblastoma-derived GSC83 and 528 cells induces the up-regulation of oncogenic splicing variants of ETV1 and NDE1 [82]. Functional SRSF3 in glioblastoma cells U-87 MG and U-118 MG is related to PDGFRB-activated PI3K-AKT-ERK pathway [83]. Oncogene HER2 produces multiple isoforms by alternative splicing. SRSF3 binds to exon 15 and inhibits the inclusion of downstream exons to increase the expression of oncogenic HER2 isoform [84].
Seeking for SRSF3-specific binding sites was initially undertaken by SELEX and identified a (A/U)C(A/U)(A/U)C motif being specific for both SRSF3 and SRSF7 [85]. Because of lacking high quality of anti-SRSF3 antibody for immunoprecipitation, various in vivo RNA-binding landscape studies of SRSF3 were undertaken by expression of a SRSF3-tagged fusion in the presence of endogenous SRSF3. Cautions should be taken in carrying out such a study because a SRSF3-tagged fusion may not truly capture the target RNAs of the endogenous SRSF3 and the excessive expression of SRSF3-tagged fusion may affect the overall gene expression profile from normal endogenous SRSF3 at a physiological or pathological condition. In mouse pluripotent embryonal carcinoma P19 cells with SRSF3-GFP expression, anti-GFP CLIP discovered hundreds of transcripts in association with SRSF3-GFP, including PTBP1, SLAIN2 and the intron-retained mRNA isoforms SRSF3 and SRSF7 [15] [86], and identified many SRSF3-GFP binding motifs with a pentamer sequence UCAUC, CUUCA, CAUCA, CAUCU, or UCAAC [87]. These pentamers are similar to the core motifs, CUCKUCY, identified by an in vitro SELEX screening assay [88]. Further studies using mouse neonatal cardiomyocytes expressing SRSF3-GFP by anti-GFP CLIP and RNA-seq showed that SRSF3 binding sites identified are mainly in the exclusion-enhanced exons, but in the flanking introns or exons for the inclusion-enhanced exons [47]. To more specifically identify targets and binding sites of SRSF3 in cells, a TRIBE (targets of RNA binding sites by editing) [89] technique by expression of SRSF3 fused with an-adenosine deaminase domain in human embryonic H9 cells was established in combination with RNA-seq analysis to identify SRSF3 target RNAs bearing A to G editing events. This technique identified 3888 confident binding sites with conserved CA-rich sequences in RNAs from 1222 genes [90]. In mouse hepatocytes, Srsf3 deletion caused insulin receptor gene (Insr) exon 11 skipping and produced isoform INSR-A in Srsf3 knockout hepatocytes, which can be strongly activated by IGF2 treatment and led to significantly enhance cell proliferation [91]. In human INSR gene, exon 11 also contains an exonic splicing enhancer of CUCUUC interacting with SRSF3 [92].
In addition to the varables derived from different assay platforms being used in the studies, the targets and binding motifs of SRSF3 identified may vary widely with different species and cell types, even though SRSF3 protein sequence is the same in both human and mouse.
4.1.2 Other splicing factors as a SRSF3 target
SRSF3 regulates not only its own exon 4 splicing but also other splicing factors' alternative splicing events. For example, MBNL1 (muscle blind-like 1) is a splicing factor with 400-aa residues and has 10 exons. Alternative exons 4 and 7 included in full-length MBNL1 is required for nuclear localization and dimerization [93]. MBNL1 inhibits the inclusion of its own exon 7. However, SRSF3 breaks its autoregulatory mechanism and enhances the inclusion of exons 4 and 7 via exonic CA-rich elements [94] [16]. Besides its own “poison” exon 4, SRSF3 also promotes the poison exons inclusion of SRSF1 [16] and SRSF7 [87] and shares many co-targets with TDP43 (TAR DNA-binding protein, also called TARDBP), an hnRNP family splicing factor [95]. For example, SRSF3 and TDP43 inhibited inclusion of SYNGAP1 exon 14 by binding to an exon 14 and an intron 15 motif, respectively, in a cooperative manner and knockdown of either SRSF3 or TDP43 expression promotes inclusion of the exon 14 [95].
4.2 SRSF3 regulation of other RNA processing steps
Other than RNA splicing regulation, SRSF3 is involved in almost all aspects of RNA biogenesis and processing including gene transcription, RNA polyadenylation, export, stability, translation, and miRNA biogenesis (Figure 3).
4.2.1 Termination of transcription
In C. elegans, depletion of Srsf3 impaired transcription termination [96]. The relative mechanism remains unclear. In mammalian cells, SRSF3 may also regulate transcriptional termination by interaction with RNA polyadenylation cleavage factor CFIm [97].
Molecular mechanisms of SRSF3 in regulation of RNA biogenesis and processing. SRSF3 regulates gene transcription, alternative splicing and polyadenylation of pre-mRNAs, RNA export, stability, and translation, and miRNA biogenesis.
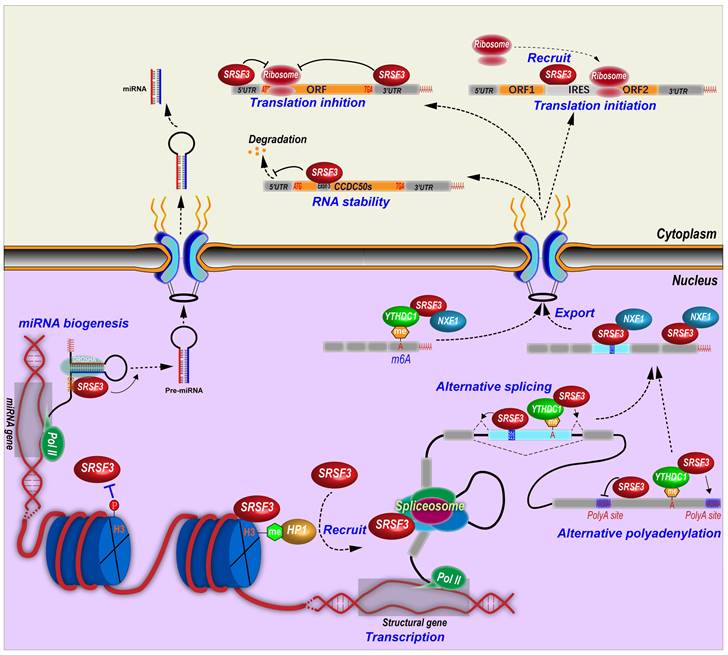
4.2.2 SRSF3 and polyadenylation
Many genes contain alternative polyadenylation sites and produces transcripts with different length of 3' untranslated regions (3' UTRs), which affects RNA stability, translation efficiency, and so on [98]. In fact, SRSF3 can regulate alternative polyadenylation in many genes. SRSF3 knockdown was found to globally promote the usage of proximal alternative polyadenylation and produce transcripts with shorten 3' UTRs, which in general facilitates protein translation, in human HEK293T cells [99]. SRSF3 binds more frequently to proximal polyadenylation sites than distal sites, and suppresses the usage of proximal sites. Notably, many genes related to cell division and cell cycle switch utilize the proximal polyadenylation sites that could be upregulated upon SRSF3 knockdown, such as PTEN, which inhibits cell proliferation and induces senescence in 293T, NIH3T3, and human umbilical vein endothelial cells [99]. Mouse Srsf3 promotes the usage of distal polyadenylation sites in mouse pluripotent P19 cells by binding to the motifs around proximal polyadenylation sites. This is because Srsf3 prevents the proximal site binding of Srsf7, which promotes the usage of proximal sites [100]. In addition, SRSF3 also regulates the selection of polyadenylation sites via an indirect mechanism. The cleavage and polyadenylation (CPA) of pre-mRNA are mainly mediated by the CPA machines including cleavage factor Im (CFIm) complex, which inhibit proximal polyadenylation site usage [101]. Mouse Srsf3 knockdown reduces Cpsf6 expression, a component of CFIm complex, due to the open reading framing shift caused by the inclusion of an alternative exon in intron 5 or the exclusion of exon 6 in Cpsf6 transcripts [100]. SRSF3 binds an intronic splicing enhancer to promote the inclusion of CT/CGRP gene exon 4 and the usage of an alternative polyadenylation site in exon 4 by interacting with cleavage stimulation factor CstF [6].
4.2.3 SRSF3 promotes RNA export
SRSF3 interacts with a 22-nt motif (ACAACAAGAAGACGCGCAUCAU) in mouse histone H2a RNA to facilitate the intronless H2a RNA export [10] and binds other histone RNAs [87]. SRSF3 interacts with the N-terminus of NXF1, the major export receptor of mRNAs [11]. SRSF3 promotes the recruitment of NXF1 to last exons of mRNAs and forms an RNA-SRSF3-NXF1 complex to be exported to the cytoplasm. Thus, SRSF3 determines the sequence specificity of exported target mRNAs [28]. This interaction between SRSF3 and NXF1 is mediated by an arginine-rich peptide near the RRM domain of SRSF3 [22].
PDCD4 (Programmed cell death 4) has a novel exon in intron 2. SRSF3 knockdown promotes the nuclear export of the transcripts with this novel exon, but has no effects on the transcripts without this novel exon [102]. PD-1 (Programmed cell death-1) mRNA nuclear export is also promoted by the interaction between SRSF3 and NXF1 [52].
NANOG is a core pluripotency transcription factor. SRSF3 facilitates mouse Nanog mRNA export and protein expression indirectly through regulation of Nxf1 RNA splicing. SRSF3 promotes Nxf1 intron 10 retention and increases a short isoform of Nxf1 protein which lacks interaction with the nuclear pore complex [103], suggesting the presence of a potential feedback regulatory mechanism by which cells can maintain relative stable level of NXF1 and RNA export levels upon SRSF3 expression.
4.2.4 SRSF3 regulates RNA stability
In the nucleus, SRSF3 plays different roles in RNA stability. SRSF3 controls the expression of the core components of NEXT (nuclear exosome targeting) complex, including RBM7, ZCCHC8 and hMTR4 [104], indicating SRSF3 is linked to a nuclear RNA decay pathway.
Mouse Srsf3 depletion increases mTOR intron 5 retention, which leads to open reading frame shift and the loss of mTOR enzyme activity. Reduced expression of mTOR full-length protein cannot maintain eIF4E-BP1 phosphorylation, and thus impairs eIF4E-BP1 binding to eIF4E, resulting in disassociation of eIF4E from the mRNA cap. Finally, mRNA decapping of contraction-related genes leads to their RNA degradation in adult mouse heart with Srsf3 knockout [47].
4.2.5 SRSF3 regulates RNA m6A modification
N6-methyladenosine (m6A) alters RNA alternative splicing. YTHDC1, a N6-methyladenosine (m6A) modified RNA binding protein or a m6A “reader”, displays similar regulatory splicing patterns as to SRSF3 but opposite to SRSF10 (SRrp40). YTHDC1 binds m6A modified exons and recruits SRSF3 to regulate exon alternative splicing, but blocks SRSF10 binding [105]. YTHDC1 regulates alternative polyadenylation. SRSF3 interacts with YTHDC1 and identifies the m6A modified sites around last exon, and then promotes the usage of distal alternative polyadenylation sites and produces a long 3'UTR region [106]. SRSF3 binds to m6A modification in ANRIL exon 1 and promotes the expression of ANRIL long isoform but inhibits the expression of ANRIL short isoform [107]. Knockdown of SRSF3 leads to decrease m6A modification level of ANRIL in pancreatic cancer cells [107]. SRSF3 facilitates the nuclear export of N6-methyladenosine (m6A) modified RNA. YTHDC1 is required for the nuclear export of m6A-containing RNA by interacting with hypo-phosphorylated SRSF3, which bridges YTHDC1 and the export receptor NXF1 [108].
4.2.6 SRSF3 in miRNA biogenesis and miRNA pathways
SRSF3 regulates the expression of miRNAs. By using miRNA microarray assays, we identified at least 20 miRNAs were regulated by SRSF3, including a subset of oncogenic or tumor suppressive miRNAs [16].
Mechanically, SRSF3 promotes miRNA biogenesis by binding to a CNNC (N represents any nucleotide) motif. This CNNC motif is around 17-18 nt downstream of the Drosha cleavage site and recruits Drosha to the basal junction and enhances the processing of the pri-miRNAs [13, 14]. This CNNC motif is correlated with a CAUC sequence interacting with the SRSF3 RRM domain [22]. More specifically, a single G to A substitution in mir-30c-1 gene facilitates SRSF3 binding and increases miR-30c-1 biogenesis and expression level in breast and gastric cancer patients [109]. Deletion of a “CNNC” motif in miR-3131 gene interferes the binding of SRSF3 and the biogenesis of mature miR-3131 [110].
SRSF3 also indirectly regulates microRNA expression. SRSF3 inhibits expression of miR-132-3p and miR-212-3p by increasing the expression of RE1-silencing transcription factor (REST), which is a transcription repressor [111]. Expression of miR-1908 is transactivated by NF-κB. SRSF3 promotes nuclear translocation of p65 (RelA), a subunit of dimeric NF-kB, to increase the expression of miR-1908 [112].
5. The roles of SRSF3 in cancer
In 2010, we discovered that SRSF3 is a proto-oncogene and overexpressed in numerous types of cancers and demonstrated that this overexpression of SRSF3 in cancer cells is critical for cancer cell proliferation and tumor induction and maintenance[12]. Following our observation, many other studies including ours have explored the expression, functions and regulatory mechanisms of SRSF3 in various cancers. Up to date, it becomes clear that SRSF3 is involved in the almost all aspects of carcinogenesis.
5.1 Expression of SRSF3 in cancers
SRSF3 overexpression is strongly positively associated with various cancers. and is overexpressed in almost all solid cancers examined, including cancers of the cervix, lung, breast, esophagus, stomach, skin, bladder, colon, liver, thyroid, and kidney, mesenchymal tissue-derived tumors, including rhadbomyosarcoma, hemangioendothelioma, hemangiopericytoma, neurofibroma, neurilemmoma, liposarcoma, leiomyosarcoma, histiocytoma, and synovial sarcoma, and B-cell lymphomas [12]. Subsequently, other studies confirmed our initial findings of SRSF3 overexpression in breast cancer [113], colon cancer [114], or cervical cancer [115], colorectal cancer [67, 116], and other types of cancers including oral squamous cell carcinoma [117], ovarian cancer [118, 119], and glioblastoma [80, 82].
In contrast to most cancers, the role of SRSF3 in hepatocellular carcinoma is complicated. Srsf3 knockout in mice appears to induce hepatocellular carcinoma [91] and overexpression of SRSF3 suppresses hepatocellular carcinoma cell proliferation. Ser/Thr phosphatase PPM1G promotes tumorigenesis of hepatocellular carcinoma cells by dephosphorylating SRSF3 [32]. However, SRSF3 mRNA levels are upregulated in hepatocellular carcinoma tissues when compared with no-tumor tissues [120]. While SRSF3 mRNA is increased, the significant reduction of SRSF3 protein, not other SR proteins, being accompanied by induction of DNA damage in hepatocellular carcinoma, appears to be related to IGF2 overexpression and activation of AKT and ERK signaling [56]. Notably, SRSF3 is essential to prevent cell apoptosis [12]. More studies are required to uncover the exact roles and expression regulatory mechanisms of SRSF3 in hepatocellular carcinogenesis.
5.2 SRSF3 regulates cellular functions associated with tumorigenesis (Figure 4)
5.2.1 Cancer cell proliferation and cell cycle progression
Malignant cells are characterized by accelerated proliferation and cell cycle progression. SRSF3 is a proto-oncogene and plays important roles in cell proliferation and cell cycle progression essential for tumorigenesis. SRSF3 overexpression promotes cell cycle progression by pushing the cells quickly through G2/M phase in synchronized mouse embryonic fibroblast cells [12]. Consistently, SRSF3 knockdown induces cell cycle arrest in G2/M stage in U2OS and HeLa cancer cells [12]. Mechanically, SRSF3 promotes expression of FOXM1, PLK1, and Cdc25B, three important cell cycle regulators [12]. SRSF3 prevents the inclusion of FOXM1 exon 9 for FOXM1a production, but enhances exon 9 exclusion to produce FOXM1b and FOXM1c, which are the key transcriptional factors for cell cycle G2/M transition [121]. As FOXM1a lacks transcriptional activities, SRSF3-mediated FOXM1b and FOXM1c transactivate PLK1 and Cdc25B expression to control cell cycle progression. Interestingly, depletion of SRSF3 seems to lead to different type of cell cycle arrest in different cell types. In HCT116 colon cancer cells, SRSF3 knockdown induces G1 cell cycle arrest due to the downregulation of cyclin D1 and E2F1, which are essential for G1-to-S cell cycle progression [114]. However, SRSF3 may also inhibit G1 phase progression by suppressing Rac family small GTPase 1 (Rac1) exon 3b splicing and the production of an oncogenic Rac1b, which plays an important role in G1 phase [122].
Cellular functions of SRSF3. SRSF3 regulates wide-range of cellular functions associated with tumorigenesis, including cell proliferation, cell cycle progression, cell apoptosis, autophagy, senescence, reprogram, and metabolism.
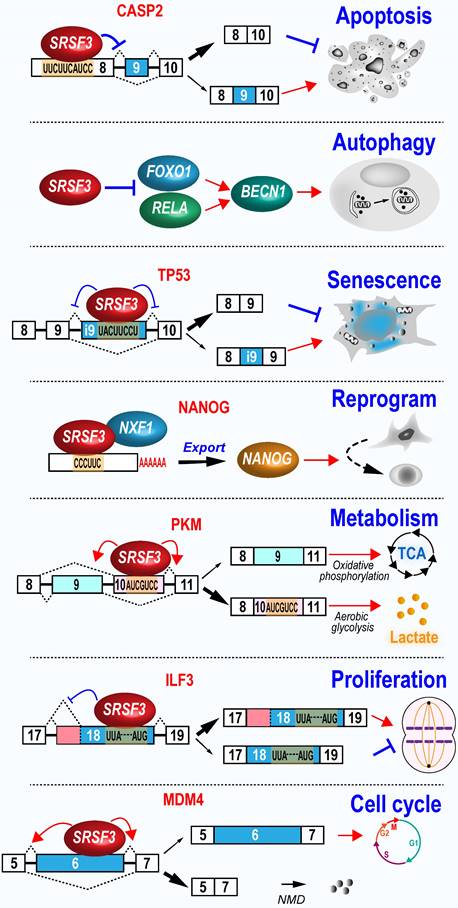
ILF3 is involved in the regulation of cell proliferation in cancer cells [123]. ILF3 gene produces multiple RNA isoforms via the alternative splicing of exon 19 to 21. The inclusion of ILF3 exon 19 leads to the usage of an alternative proximal polyadenylation site. Moreover, ILF3 exon 20 and 21 are two alternative exons. We found that SRSF3 increases exon 19 inclusion and the usage of a proximal 3' splice sit to produce oncogenic ILF3 isoform-2, as well as the inclusion of exon 20 and 21 to produce oncogenic ILF3 isoform-1, by interacting with several motifs in exon 18. In contrast, tumor suppressive ILF3 isoform-5 and -7 expression were inhibited by SRSF3 [80].
MDM4 protein inactivates p53 function and is often upregulated in highly proliferating stem cells or cancer cells. The increased MDM4 protein level in these cells is due to increased translational activities as its mRNA level often remains unchanged. Mechanically, MDM4 exon 6 is an alternative exon. Exclusion of exon 6 disrupts its open reading frame to produced an unstable short RNA isoform as a NMD target. SRSF3 promotes exon 6 inclusion and full-length MDM4 protein expression. An oligonucleotide targeting the SRSF3 binding sites in exon 6 represses exon 6 inclusion and MDM4 protein expression, and thereby, inhibits the development of melanoma and diffuse large B cell lymphoma [124].
Glioblastoma is one of the most aggressive brain tumors characterized by fast growth and spread, and chemotherapy resistance. SRSF3 is required for glioblastoma cancer cell proliferation and tumorigenesis by promoting exon 7 inclusion and protein stability of transcription factor ETV1 (ETS variant 1), which enhances cell proliferation and invasion[82].
5.2.2 SRSF3 inhibits apoptosis
Increased SRSF3 expression is necessary for indefinite growth of cancer cells and prevents cancer cell apoptosis. In contrast, depletion of SRSF3 in many types of cancer cells examined inhibits cell proliferation and induce cell apoptosis initially found in U2OS cells [12]. Subsequently, a series of studies have revealed that SRSF3 knockdown induces significant high level of caspase-3 cleavage, condensed and fragmented nuclei, and cell proliferation inhibition in U2OS, SW480 (a human colon adenocarcinoma cell line) [125], and ovarian cancer cells [119]. HIPK2 (Homeodomain-interacting protein kinase-2) induces apoptosis. SRSF3 regulates alternative splicing of HIPK2 exon 8 to prevent HIPK2-related apoptosis [114], but increases the production of anti-apoptotic protein BCL2 in colon cancer cells [114].
On the other hand, SRSF3 has a role in anti-apoptosis and promotes exon 9 skipping of caspase-2 [126]. Caspase-2 (CASP2) exon 9 is alternative exon. Exon 9 inclusion produces pro-apoptotic caspase-2L isoform, while exon 9 exclusion produces anti-apoptotic caspase-2S isoform.
5.2.3 SRSF3 protects cells from senescence
Srsf3 expression is significantly reduced in spleen and muscle of old mice compared with young mice [127]. It is not surprising that SRSF3 may have an anti-aging function, because reduced SRSF3 expression leads to cellular senescence in normal human fibroblast cell line [48], human umbilical vein endothelial cells, and mouse fibroblast cell line [99]. SRSF3 protects cell from senescence. In normal human fibroblasts, SRSF3 knockdown induces cellular senescence by increasing p53 (TP53) phosphorylation at serine 15, and production of p53β isoform, which can promotes p53-mediated senescence [48]. Human fibroblasts undergoing replicative senescence exhibit reduced expression of SRSF3. SRSF3 knockdown also increased p53β expression and senescence in neural astrocytes [128]. In principle, SRSF3 promotes the inclusion of a p53β-specific exon via interacting with three motifs, UUUCAAA, UACUUGAC, and UACUUCCU [48]. SRSF3 prevents the usage of proximal polyadenylation sites in many genes associated with senescence pathways, such as PTEN and inhibits the expression of these genes in senescence-related phenotypes [99]. Cells undergoing senescence remain viable but fail to proliferate. Therefore, senescence can suppress tumorigenesis by blocking uncontrolled cell growth in cancer cells [129]. Thus, SRSF3 is an inhibitor of cellular senescence and promotes cell growth.
5.2.4 SRSF3 represses autophagy
Autophagy helps cells to recycle materials and is essential in physiological and pathological conditions. SRSF3 is significantly downregulated during hypoxia-induced autophagy. Knockdown of SRSF3 induces significant cellular autophagic activity, whereas SRSF3 overexpression inhibits autophagy in oral squamous cell carcinoma cells. Mechanically, SRSF3 inhibits the expression of p65 (RELA) and FOXO1 to reduce expression of BECN1, a key regulator of autophagy [130].
5.2.5 SRSF3 is required for the homologous recombination-mediated DNA repair
In ovary cancer cells, SRSF3 inhibits DNA double-strand break, and promotes homologous recombination-mediated DNA repair. This is because SRSF3 regulates the expression of the genes involved in DNA double-strand break repair. Knockdown of SRSF3 expression decreases the expression of BRCA1, BRIP1, and RAD51, which are responsible for homologous recombination-mediated DNA repair, at both transcription and protein levels, but has no effect on RNA splicing of these genes [131]. In pancreatic cancer, SRSF3 increases the expression of lncRNA ANRIL long isoform by recognizing its m6A modification. The long isoform of lncRNA ANRIL promotes DNA homologous recombination repair by binding to DNA damage sites to recruit DNA repair proteins RAD50, BRCA1, BRCA2, and RAD51 [107]. Knockout of mouse Srsf3 in hepatocytes causes DNA damage and the decreased expression of Xrcc1 (X-ray repair cross-complementing protein 1), Msh2 (mutS homolog 2), and Xpd (xeroderma pigmentosum group D) [56].
5.2.6 SRSF3 enhances cell reprogram
During the reprograming progress in mouse embryonic fibroblast, Srsf3 expression level undergoes a relative lower increase, followed by a sharp increase, which is correlated with the increases of cell proliferation genes and activation of genes of stem cell maintenance [103]. Overexpression of Srsf3 in mouse embryonic fibroblast significantly enhances the reprogramming by binding Nanog mRNA and promoting its nuclear export and protein expression [103]. Moreover, increased expression of mouse Srsf3 has been noted in induced pluripotent stem cells (iPSCs) when compared with parent somatic cells. Knockdown of mouse Srsf3 displays no effect on somatic cell proliferation, but significantly suppresses the efficiency of somatic cell reprogramming [132].
5.2.7 SRSF3 regulates metabolism
PKM (Pyruvate kinase M) gene produces two isoforms, M1 and M2, via mutual splicing of exon 9 or 10 [133]. M1 promotes oxidative phosphorylation and is mainly expressed in normal muscle and brain tissues, while M2 promotes aerobic glycolysis and is mainly expressed in embryonic and cancer tissues [134]. A potent SRSF3-binding exonic splicing enhancer in exon 10 is responsible for M2 expression. Knockdown of SRSF3 significantly decreases lactate production and cancer cell proliferation by decreasing exon 10 inclusion which is independently of exon 9 inclusion [135]. In contrast, knockdown of SRSF3 increases M1 protein level and thus, the ratio of M1 vs M2 protein levels, to induce the energy metabolic shift from glycolysis and oxidative phosphorylation in colorectal cancer cells [116]. This SRSF3/PKM/glycolytic pathway also happens in dendritic cells [136].
Natural and synthetic glucocorticoids bind GR (glucocorticoid receptor) to maintain various cellular metabolic and homeostatic functions [137]. GR contains two isoforms generated by alternative splicing of exon 9 of NR3C1 gene. The usage of proximal 5' splice site of exon 9 produces GRα, which is the classic receptor for glucocorticoid receptor signaling. Whereas the usage of distal 5' splice site of exon 9 produces GRβ, which plays the dominant negative effects and is upregulated in a variety of glucocorticoid resistant diseases [137]. SRSF3 promotes the usage of proximal splice site of exon 9 and the product of GRα in breast cancer cells [138] or THP-1 cells [139]. However, SRSF3 in HeLa cell does not regulate GR exon 9 splicing [140], indicating that the effects of SRSF3 on GR exon 9 splicing is cell type dependent. Interestingly, SRSF3 enhances glucocorticoid receptor signaling by increasing GRα expression and is upregulated upon cortisol stimulation[138].
5.2.8 SRSF3 and stress responses
Stress granules are membraneless cytoplasmic RNA particles containing stalled mRNA-protein complexes in response to various stresses [141]. SRSF3 is a component of stress granules and is required for its assembly, indicating its important role in mRNA metabolism in the cytoplasm [142]. Under arsenite-induced oxidative stress, SRSF3 is neddylated in Lys85 and promotes stress granule aggregation in human osteosarcoma U2OS cells [26]. However, SRSF3 in hepatocytes is neddylated at Lys11 and degraded by proteasome under palmitic acid-induced oxidative stress [25]. These results suggests that the distribution, function, and fate of SRSF3 in different cells are regulated possibly by the neddylation in its different Lys sites under various stress conditions.
5.3 SRSF3 promotes tumorigenesis (Figure 5)
5.3.1 SRSF3 in malignant transformation progress
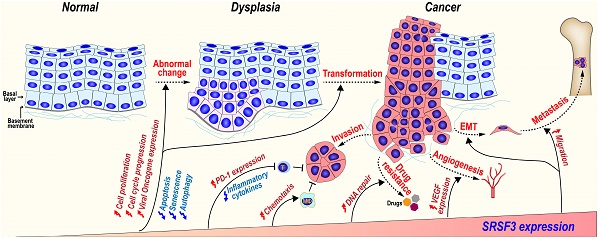
SRSF3 expression is gradually increased during the malignant transformation progress from normal oral mucosal, dysplasia (precancerous lesions), to oral squamous cell carcinoma [117]. In a mouse model of mammary tumorigenesis, Srsf3 expression is dramatically up-regulated during the progress of mammary cancer in both mRNA and protein levels [143]. We found that SRSF3 in overexpression could transform mouse embryonic fibroblast and induce tumors in nude mice via upregulating the expression of FoxM1 and its downstream targets Cdc25B and PLK1 [12], as well as promoting the expression of ILF3 isoform-1 and isoform-2 [80]. These results suggest SRSF3 play an important role in malignant transformation progression.
5.3.2 SRSF3 in cancer invasion and metastasis
SRSF3 may promote cancer invasion and metastasis via several mechanisms.
MAP4K4 (Mitogen-activated protein kinase kinase kinase kinase 4) regulates cell transformation and invasion [144]. We found SRSF3 is required for the inclusion of MAP4K4 novel exon 23alt [16]. SRSF3 also promotes the inclusion of MAP4K4 exon 16 via CU rich elements to induce the expression of epithelial mesenchymal transition (EMT)-related genes [67].
SRSF3 plays important roles during tumorigenesis. SRSF3 is overexpressed in dysplasia and cancer tissues. SRSF3 in overexpression promotes cell malignant transformation and the invasion, metastasis, angiogenesis, drug resistance, and immunosuppression of cancers.
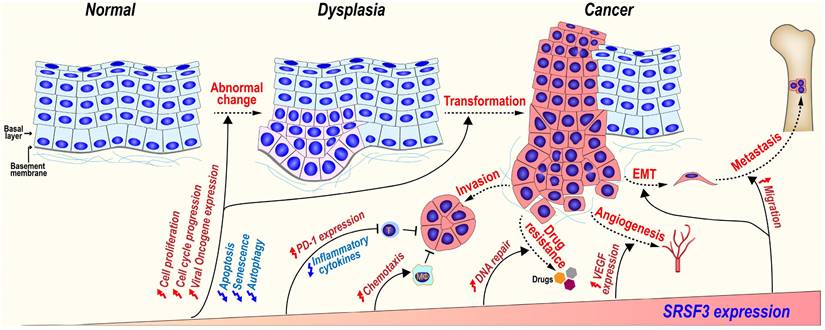
CD44 is a cell surface hyaluronan-binding glycoprotein and a stem cell biomarker, which enhances cell migration by mediating cell-cell and cell-extracellular matrix interactions [145]. SRSF3 increases the inclusion of exon v8-v10 of CD44 to produce oncogenic isoform CD44E in gastric cancer [146] and promotes the inclusion of variable exons of CD44 isoforms in triple-negative breast cancer (TNBC) [147]. SRSF3 recognizes an exonic splicing enhancer (5'-CUUCGAUCAACGCCACGCCA-3') in CD44 exon v9 [45].
SRSF3 can also promote metastasis and anoikis resistance by enhancing the inclusion of CPEB2 exon 4 via binding to a “CAUCC” splicing enhancer, and producing full-length CPEB2B protein to activate the translation of TWIST1 and HIF1α in triple negative breast cancer [113]. In a mouse breast cancer cell line, Srsf3 knockdown significantly reduces cellular migration, invasion in vitro, and lung metastasis in vivo. Mechanically, SRSF3 forms a complex with TDP43 and promotes the exclusion of PAR3 (partitioning defective protein 3) exon 12 to produce an oncogenic isoform in coordination with splicing factor TDP43, while full-length PAR3 is a metastasis inhibitor [95].
5.3.3 SRSF3 and cancer drug resistance
Gemcitabine, a widely used chemotherapy drug, kills cancer cells by inhibiting DNA replication and cell division. However, cancer cells often develop resistance to gemcitabine [148]. SRSF3 is highly expressed in gemcitabine-resistant pancreatic cancer cells. The increased expression of SRSF3 enhances gemcitabine resistance of pancreatic cancer cells both in vitro and in vivo, whereas silence of SRSF3 by siRNAs exhibits an opposite effect.
5.3.4 SRSF3 promotes angiogenesis
Angiogenesis is essential for the development of cancer. Knockdown of SRSF3 expression significantly inhibits VEGF (vascular endothelial growth factor) expression in colorectal cancer cells, cancer cell migration, and tube formation by human umbilical vein endothelial cells (HUVECs) in vitro via a proangiogenic SRF (serum response factor) gene. SRSF3 is co-expressed with SRF in colorectal cancer tissues, and required for SRF expression [149].
5.4 SRSF3 is a negative regulator of immune response
5.4.1 SRSF3 in inflammation and innate immune responses
SRSF3 regulates innate immune responses. SRSF3 inhibits IL-1β expression in a monoclonal cell line, THP-1 cells, upon the challenge of Escherichia coli. Bacterial treatment significantly reduces SRSF3 expression to allow IL-1β expression in primary monocytes [150]. In dendritic cells, histone deacetylase (HDAC) inhibitor TSA upregulates SRSF3 and downregulates pro-inflammatory cytokines, including IL-1β, IL-10, and IL-12 [136].
A set of innate immune associated genes, such as Saa3, Ccl5, Ccl3, and Lcn2, are highly transcribed, but not efficiently translated under inflammation stimulation in brain microglial cells. Srsf3 binds to the 3' UTR of these genes and represses their translation. Interestingly, Srsf3 expression is upregulated by LPS-activated mouse brain microglial cells and may suppress uncontrolled inflammation responses and tissue damage [151].
5.4.2 SRSF3 and cancer immunotherapy
SRSF3 is involved in regulating the expression of genes related to cancer immunotherapy. PD-1 (Programmed cell death-1) is an important immunosuppressor molecule for cancer immunotherapy [152]. TGF-β1 induced Smad3 to transactivate PD-1 expression in T cells to restrict their antitumor function [153]. Besides Smad3, TGF-β1 also promotes PD-1 expression by two mechanisms mediated by SRSF3. TGF-β1 promotes SRSF3 expression. Upregulated SRSF3 binds to and prolongs PD-1 mRNA half-life by increasing its stability, and promotes its nuclear export by the interaction with NXF1 [52].
In B-cell acute lymphoblastic leukemias (B-ALL), SRSF3 expression seems to be beneficial for patients. CD19 is a target antigen in B-ALL. Mutation or skipping of CD19 exon 2 induces the resistance to chimeric antigen receptor-armed T cell (CART-19) treatment [154]. SRSF3 is required for the inclusion of exon 2 by interacting with the [A/T]CAAC[A/G] motifs, and is downregulated in relapsed leukemias after immunotherapy [154].
Roles of SRSF3 in anti-tumor immune response can be exemplified by YAP1 (Yes-associated protein 1), an oncogenic co-transactivator. YAP1-2γ contains a γ fragment encoded by an alternative exon 6 and exhibits significant anti-tumor activity by enhancing CCL2 expression and attracting massive macrophage infiltration. SRSF3 promotes exon 6 inclusion and results in the upregulation of CCL2. Oncogenic KRAS downregulates SRSF3 expression, YAP-1 exon 6 inclusion, and thus, isoform YAP1-2γ expression [155]. Altogether, although SRSF3 negatively regulates inflammation and innate immune responses, SRSF3 may play double-edged roles in cancer immunotherapy.
5.5 Oncogenic viruses take advantage of SRSF3
Persistent infections with high-risk human papillomaviruses (HPV) lead to development of cancers. Almost all cervical cancer is caused by high-risk HPV. We discovered that SRSF3 interacted with a HPV16 A/C-rich ESE motif (nt 3488 to 3516), by which SRSF3 inhibits HPV16 later gene expression, but increases the expression of viral early genes. In cervix epithelium, SRSF3 expression gradually decreases from the undifferentiated basal layers to the highly differentiated upper layers. The viral late genes, L1 and L2, can only be expressed in the upper layers within the keratinocytes with reduced SRSF3 expression [34]. Similarly, we also discovered an ESE motif (nt 3520 to 3550) in HPV18, through which SRSF3 suppressed HPV18 L1 and E2 expression [62]. Importantly, SRSF3 is required for the expression of both HPV18 and HPV16 early and oncogenic genes, E6 and E7, in HPV18-positive HeLa and HPV16-positive CaSki cells, two commonly used cervical cancer cell lines [34].
Kaposi sarcoma-associated herpesvirus (KSHV) RNA-binding protein ORF57 (Mta) functions as a viral splicing factor and a key post-transcriptional regulator in viral gene expression [156] . SRSF3 inhibits KSHV K8β splicing by binding to a suboptimal K8β intron. The N-terminal ORF57 binds the SRSF3 RRM, prevents SRSF3 association with the K8β intron, and promotes K8β splicing [157]. Moreover, other targets of SRSF3 were also reversely regulated by ORF57, which suggested that ORF57 might regulate cellular gene splicing via SRSF3 during KSHV infection [157].
SRSF3 is also used by other viruses for replication. During poliovirus infection in neuroblastoma cells, SRSF3 migrates from the nucleus to the cytoplasm, and co-localizes to poliovirus RNA with PCBP2 via its RS domain [158]. PCBP2 binds to the major stem-loop structure (stem-loop IV) in the IRES of poliovirus RNA and facilitates the translation initiation[159, 160]. Poliovirus reduces cellular translation, but translates its own RNA by the IRES (internal ribosome entry sites)-mediated translation, which is enhanced by recruiting PCBP2 and SRSF3 [158, 161].
In herpes simplex virus (HSV-1), viral mRNA export is mainly mediated by HSV-1 ICP27 protein. However, SRSF3 can assist the export of ICP27-mediated viral mRNA export. Depletion of SRSF3 led to about a 10-fold decrease in virus yield [162]
In oncogenic Epstein-Barr virus associated with development of Burkitt lymphoma, nasopharyngeal carcinoma, and Hodgkin lymphoma, viral EB2 protein (also called SM, Mta, and BMLF1), a KSHV ORF57 homolog, interacts with the RS domain of SRSF3, and activates a cryptic alternative 5' splice site in intron 23 of STAT1 to produce a dominant negative repressor of STAT1 protein [163]. STAT1 is a mediator of interferon (IFN) signal pathway. Thus, EBV EB2 protein uses SRSF3 to disrupt IFN signal pathway. SRSF3 destablizes viral intronless RNAs by interacting with nuclear RNA exosome and its adaptor complex NEXT in the absence of EBV EB2, but this effect could be counteracted by viral EB2 via interacting with SRSF3 [104].
6. SRSF3 functions in other human diseases
6.1 SRSF3 and cardiac diseases
Mouse Srsf3 is essential for both heart development and function. Depletion of Srsf3 specifically in embryonic heart causes failure of cardiomyocyte proliferation, while depletion of Srsf3 in cardiomyocytes of adult mice causes severe systolic dysfunction and death [47]. Srsf3 depletion causes mitochondrial matrix swelling and disorganized cristae in cardiomyocytes, accompanied by a significant reduction in complex I-driven respiration due to the decreased mitochondrial DNA and expression of oxidative phosphorylation-related genes, such as Ndufb8, Sdhb, and Mtco1, and aberrant alternative splicing of mitochondrial genes, such as Bag3 and Atg13 [164].
6.2 SRSF3 and neurological diseases
SRSF3 is overexpressed in Alzheimer's disease (AD) neuronal cells, which is attributed to the pathological Aβ42 fibril stimulation [165]. Increased SRSF3 promotes the inclusion of alternative exon 19 of TrkB (tropomyosin receptor kinase B) receptor, which mediates the signal transduction of BDNF (brain-derived neurotrophic factor) to support neuronal survival, growth, and differentiation. The inclusion of TrkB alternative exon 19 leads to encoding a truncated TrkB protein lacking the tyrosine kinase domain. Truncated TrkB protein interferes the BDNF signal transduction and contributes to AD [165].
Wnt1-Cre conditional knockout of mouse Srsf3 led to facial cleft in mouse due to the defective proliferation of cranial neural crest cells and the dysregulated alternative splicing of some protein kinases in PDGFRα (platelet-derived growth factor receptor alpha) signaling pathway, such as Limk2, Dmpk and Prkd2 genes [30].
6.3 SRSF3 and development
Megakaryocyte specific knockout of mouse Srsf3 blocks the maturation of megakaryocytes and the deposition of distinct megakaryocyte RNAs into platelets, leading to dramatically decreased platelet amount and dysfunctional platelets [166].
Srsf3 is essential for mouse liver development. Hepatic specific knockout of Srsf3 often results in early neonatal and perinatal death with significantly smaller body size than wild-type mice. The survived Srsf3 knockout mice also shows a significant smaller body size and liver, growth retardation, and disturbed hepatic architecture [167] because Srsf3 knockout impairs hepatocyte maturation by induction of aberrant RNA splicing of the genes involved in the regulation of glucose and lipid metabolism, such as Hnf1α, Ern1, Hmgcs1, Dhcr7 and Scap [167]. G6PD (Glucose-6-phosphate dehydrogenase) is an enzyme involved in lipogenesis in liver. Starvation induces retention of the introns 11 and 12 around exon 12 of G6PD to reduce G6PD expression. Conversely, nutrients stimulates splicing of the introns and to produce G6PD protein [168]. There is a regulatory element in the exon 12 for its splicing [169]. SRSF3 binds this element and promotes exon 12 inclusion and splicing of the introns 11 and 12, and thus, helps the cells to produce more G6PD protein upon nutrient [54]. HnRNP K, hnRNP L, and hnRNP A2/B1 bind the element and suppress exon 12 inclusion [170, 171].
7. Current progress in developing chemical inhibitors to control SRSF3 expression
SRSF3 overexpression is partially resulted from the reduced inclusion of its poison exon 4 and the impaired autoregulation in cancers [21]. An antisense oligonucleotide blocking an exonic splicing suppressor in the exon 4 can significantly increase inclusion of the exon 4, leading to inhibit SRSF3 expression and suppress proliferation of oral squamous cell carcinoma [172] and breast cancer cells [173].
Recently, a novel SRSF3 inhibitor, namely SFI003, has been developed. By screening compounds from commercial databases, SFI003, among dozens of its derivatives, has been found to be the most potent inhibitor on SRSF3 activities, with a lowest IC50 value, in colorectal cancer cells. Moreover, SFI003 induces the neddylation-dependent degradation of SRSF3 protein and promotes apoptosis, thereby inhibiting cell proliferation, migration, and tumor formation [174].
Other chemicals also have the abilities to reduce SRSF3 expression. Amiloride, a diuretic, induces hypo-phosphorylation and downregulation of SRSF3 and SRSF1, leading to alteration of RNA splicing patterns in many genes and cell cycle disruption in hepatocellular carcinoma cells [175].
Digitoxin, a structural analogue of digoxin, also regulates a large set of alternative RNA splicing events by repressing SRSF3 expression [176]. Mechanically, digoxin decreases SRSF3 expression by inducing its protein ubiquitination and degradation. Conversely, DARPP-32 (dopamine and cAMP-regulated phosphoprotein of molecular weight 32 kDa), a key signaling mediator in several neurotransmitter pathways [177], encoded by PPP1R1B gene, binds SRSF3 protein and decreases digitoxin-induced ubiquitination of SRSF3 protein, and thus significantly increases SRSF3 stability [146].
8. Summary
SRSF3 protein and its alternative exon 4 are well conserved during evolution from animal kingdom. As a multifunctional RNA-binding protein and splicing factor, auto- and balanced regulatory mechanisms in SRSF3 expression in host cells are essential in maintaining the proper expression level of SRSF3 protein, which is often disrupted in cancers. SRSF3 is involved in many cellular processes by regulating almost all aspects of RNA biogenesis and processing of many target genes. Thus, any disturbance of SRSF3 expression and modification will contribute to turbulence of normal cell functions and the pathological outcomes. As a proto-oncogene, overexpression of SRSF3 has proven to induce cell transformation and tumor formation and maintenance. Conceivably, all types of cancers examined to date exhibit increased expression of SRSF3, which is essential for keeping cancer cell proliferation and cell cycle progression. Deficiency of SRSF3 also causes severe diseases by deregulation of RNA processing of many target genes. Although restoring a proper expression level of SRSF3 in specific cells may be the key for curing SRSF3 deficiency-induced diseases, blocking SRSF3 expression and its functions in cancer cells by small molecules will be an exciting and promising roadmap for future cancer therapies.
Supplementary Material
Supplementary figures.
Acknowledgements
This study was supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research (ZIASC010357 to Z.M.Z.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115(3):587-596
2. Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6(5):837-847
3. Ayane M, Preuss U, Kohler G, Nielsen PJ. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding motifs. Nucleic Acids Res. 1991;19(6):1273-1278
4. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24(11):1073-1074
5. Zahler AM, Neugebauer KM, Stolk JA, Roth MB. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13(7):4023-4028
6. Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18(9):4977-4985
7. Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11(9):1148-1159
8. de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13(11):973-980
9. Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9(16):899-902
10. Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7(4):899-905
11. Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11(3):837-843
12. Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6(7):806-826
13. Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152(4):844-858
14. Kim K, Nguyen TD, Li S, Nguyen TA. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA. 2018;24(7):892-898
15. Anko ML, Morales L, Henry I, Beyer A, Neugebauer KM. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat Struct Mol Biol. 2010;17(8):962-970
16. Ajiro M, Jia R, Yang Y, Zhu J, Zheng ZM. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 2016;44(4):1854-1870
17. Yu LL, Majerciak V, Jia R, Zheng ZM. Revisiting and corrections to the annotated SRSF3 (SRp20) gene structure and RefSeq sequence from the human and mouse genomes. Cell Insight. 2023;2(2):100089
18. Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17(6):3116-3124
19. Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446(7138):926-929
20. Garcia-Moreno JF, Romao L. Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay. Int J Mol Sci. 2020;21(24):9424
21. Guo J, Jia J, Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep. 2015;5:14548
22. Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J. et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25(21):5126-5137
23. Kano S, Nishida K, Kurebe H, Nishiyama C, Kita K, Akaike Y. et al. Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am J Physiol Cell Physiol. 2014;306(3):C250-262
24. Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325-340
25. Kumar D, Das M, Sauceda C, Ellies LG, Kuo K, Parwal P. et al. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J Clin Invest. 2019;129(10):4477-4491
26. Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang GY, Baek JH. et al. NEDDylation promotes stress granule assembly. Nat Commun. 2016;7:12125
27. Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci U S A. 2004;101(26):9666-9670
28. Muller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC. et al. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30(5):553-566
29. Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399(3):333-338
30. Dennison BJC, Larson ED, Fu R, Mo J, Fantauzzo KA. Srsf3 mediates alternative RNA splicing downstream of PDGFRalpha signaling in the facial mesenchyme. Development. 2021;148(14):dev199448
31. Long Y, Sou WH, Yung KWY, Liu H, Wan SWC, Li Q. et al. Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J Biol Chem. 2019;294(4):1312-1327
32. Chen D, Zhao Z, Chen L, Li Q, Zou J, Liu S. PPM1G promotes the progression of hepatocellular carcinoma via phosphorylation regulation of alternative splicing protein SRSF3. Cell Death Dis. 2021;12(8):722
33. Murray MV, Kobayashi R, Krainer AR. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999;13(1):87-97
34. Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C. et al. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009;83(1):167-180
35. Yu L, Zheng ZM. Human Papillomavirus Type 16 Circular RNA Is Barely Detectable for the Claimed Biological Activity. mBio 2022:e0359421.
36. Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12(1):55-66
37. Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A. 2001;98(18):10154-10159
38. Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138(2):225-238
39. Fitzgerald KD, Chase AJ, Cathcart AL, Tran GP, Semler BL. Viral proteinase requirements for the nucleocytoplasmic relocalization of cellular splicing factor SRp20 during picornavirus infections. J Virol. 2013;87(5):2390-2400
40. Dumont AA, Dumont L, Berthiaume J, Auger-Messier M. p38alpha MAPK proximity assay reveals a regulatory mechanism of alternative splicing in cardiomyocytes. Biochim Biophys Acta Mol Cell Res. 2019;1866(12):118557
41. Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T. et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33(4):450-461
42. Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP. et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10(7):1122-1134
43. Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256-1269
44. Baker M, Petasny M, Taqatqa N, Bentata M, Kay G, Engal E. et al. KDM3A regulates alternative splicing of cell-cycle genes following DNA damage. RNA. 2021;27(11):1353-1362
45. Galiana-Arnoux D, Lejeune F, Gesnel MC, Stevenin J, Breathnach R, Del Gatto-Konczak F. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J Biol Chem. 2003;278(35):32943-32953
46. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397-406
47. Ortiz-Sanchez P, Villalba-Orero M, Lopez-Olaneta MM, Larrasa-Alonso J, Sanchez-Cabo F, Marti-Gomez C. et al. Loss of SRSF3 in Cardiomyocytes Leads to Decapping of Contraction-Related mRNAs and Severe Systolic Dysfunction. Circ Res. 2019;125(2):170-183
48. Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM. et al. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene. 2013;32(22):2792-2798
49. Jamieson C, Sharma M, Henderson BR. Targeting the beta-catenin nuclear transport pathway in cancer. Semin Cancer Biol. 2014;27:20-29
50. Goncalves V, Matos P, Jordan P. The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14(12):2538-2549
51. Corbo C, Orru S, Gemei M, Noto RD, Mirabelli P, Imperlini E. et al. Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumorigenicity. Proteomics. 2012;12(12):2045-2059
52. Wu P, Geng B, Chen Q, Zhao E, Liu J, Sun C. et al. Tumor Cell-Derived TGFbeta1 Attenuates Antitumor Immune Activity of T Cells via Regulation of PD-1 mRNA. Cancer Immunol Res. 2020;8(12):1470-1484
53. Munoz U, Puche JE, Hannivoort R, Lang UE, Cohen-Naftaly M, Friedman SL. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol Cancer Res. 2012;10(9):1216-1227
54. Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Serine arginine splicing factor 3 is involved in enhanced splicing of glucose-6-phosphate dehydrogenase RNA in response to nutrients and hormones in liver. J Biol Chem. 2013;288(4):2816-2828
55. Saeki K, Yasugi E, Okuma E, Breit SN, Nakamura M, Toda T. et al. Proteomic analysis on insulin signaling in human hematopoietic cells: identification of CLIC1 and SRp20 as novel downstream effectors of insulin. Am J Physiol Endocrinol Metab. 2005;289(3):E419-428
56. Kumar D, Das M, Oberg A, Sahoo D, Wu P, Sauceda C. et al. Hepatocyte Deletion of IGF2 Prevents DNA Damage and Tumor Formation in Hepatocellular Carcinoma. Adv Sci (Weinh) 2022:e2105120.
57. Faenza I, Matteucci A, Manzoli L, Billi AM, Aluigi M, Peruzzi D. et al. A role for nuclear phospholipase Cbeta 1 in cell cycle control. J Biol Chem. 2000;275(39):30520-30524
58. Bavelloni A, Faenza I, Cioffi G, Piazzi M, Parisi D, Matic I. et al. Proteomic-based analysis of nuclear signaling: PLCbeta1 affects the expression of the splicing factor SRp20 in Friend erythroleukemia cells. Proteomics. 2006;6(21):5725-5734
59. Brady LK, Wang H, Radens CM, Bi Y, Radovich M, Maity A. et al. Transcriptome analysis of hypoxic cancer cells uncovers intron retention in EIF2B5 as a mechanism to inhibit translation. PLoS Biol. 2017;15(9):e2002623
60. Mole S, McFarlane M, Chuen-Im T, Milligan SG, Millan D, Graham SV. RNA splicing factors regulated by HPV16 during cervical tumour progression. J Pathol. 2009;219(3):383-391
61. Klymenko T, Hernandez-Lopez H, MacDonald AI, Bodily JM, Graham SV. Human Papillomavirus E2 Regulates SRSF3 (SRp20) To Promote Capsid Protein Expression in Infected Differentiated Keratinocytes. J Virol. 2016;90(10):5047-5058
62. Ajiro M, Tang S, Doorbar J, Zheng ZM. Serine/Arginine-Rich Splicing Factor 3 and Heterogeneous Nuclear Ribonucleoprotein A1 Regulate Alternative RNA Splicing and Gene Expression of Human Papillomavirus 18 through Two Functionally Distinguishable cis Elements. J Virol. 2016;90(20):9138-9152
63. Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16(16):5077-5085
64. Jumaa H, Nielsen PJ. Regulation of SRp20 exon 4 splicing. Biochim Biophys Acta. 2000;1494(1-2):137-143
65. Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F. et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19(2):480
66. Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q. et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374-389
67. Lin JC, Lee YC, Tan TH, Liang YC, Chuang HC, Fann YC. et al. RBM4-SRSF3-MAP4K4 splicing cascade modulates the metastatic signature of colorectal cancer cell. Biochim Biophys Acta Mol Cell Res. 2018;1865(2):259-272
68. Elizalde M, Urtasun R, Azkona M, Latasa MU, Goni S, Garcia-Irigoyen O. et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124(7):2909-2920
69. Jimenez M, Urtasun R, Elizalde M, Azkona M, Latasa MU, Uriarte I. et al. Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47(7):3450-3466
70. Jia R, Zhang S, Liu M, Zhang Y, Liu Y, Fan M. et al. HnRNP L is important for the expression of oncogene SRSF3 and oncogenic potential of oral squamous cell carcinoma cells. Sci Rep. 2016;6:35976
71. Lu GY, Liu ST, Huang SM, Chang YL, Lin WS. Multiple effects of digoxin on subsets of cancer-associated genes through the alternative splicing pathway. Biochimie. 2014;106:131-139
72. Jeandet P, Sobarzo-Sanchez E, Uddin MS, Bru R, Clement C, Jacquard C. et al. Resveratrol and cyclodextrins, an easy alliance: Applications in nanomedicine, green chemistry and biotechnology. Biotechnol Adv. 2021;53:107844
73. Markus MA, Marques FZ, Morris BJ. Resveratrol, by modulating RNA processing factor levels, can influence the alternative splicing of pre-mRNAs. PLoS One. 2011;6(12):e28926
74. Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K. et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12(5):568-573
75. Zhang Z, Huang L, Zhao W, Rigas B. Annexin 1 induced by anti-inflammatory drugs binds to NF-kappaB and inhibits its activation: anticancer effects in vitro and in vivo. Cancer Res. 2010;70(6):2379-2388
76. Gordinier ME, Zhang HZ, Patenia R, Levy LB, Atkinson EN, Nash MA. et al. Quantitative analysis of transforming growth factor beta 1 and 2 in ovarian carcinoma. Clin Cancer Res. 1999;5(9):2498-2505
77. Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23(24):4225-4231
78. Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K. et al. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A. 2006;103(10):3775-3780
79. Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14(3):185-193
80. Jia R, Ajiro M, Yu L, McCoy P Jr, Zheng ZM. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA. 2019;25(5):630-644
81. Do DV, Strauss B, Cukuroglu E, Macaulay I, Wee KB, Hu TX. et al. SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements. Cell Discov. 2018;4:33
82. Song X, Wan X, Huang T, Zeng C, Sastry N, Wu B. et al. SRSF3-Regulated RNA Alternative Splicing Promotes Glioblastoma Tumorigenicity by Affecting Multiple Cellular Processes. Cancer Res. 2019;79(20):5288-5301
83. Fuentes-Fayos AC, Vazquez-Borrego MC, Jimenez-Vacas JM, Bejarano L, Pedraza-Arevalo S, F LL. et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness: oncogenic role of SRSF3. Brain. 2020;143(11):3273-3293
84. Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12(10):1139-1151
85. Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5(3):468-483
86. Brugiolo M, Botti V, Liu N, Muller-McNicoll M, Neugebauer KM. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res. 2017;45(18):10452-10465
87. Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I. et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13(3):R17
88. Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19(3):1705-1719
89. McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W. et al. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell. 2016;165(3):742-753
90. Jin S, Xue Z, Zhang J, Wang Z, Zhang J, Chen D. et al. Identification of SRSF3 target mRNAs using inducible TRIBE. Biochem Biophys Res Commun. 2021;578:21-27
91. Sen S, Langiewicz M, Jumaa H, Webster NJ. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61(1):171-183
92. Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29(3):871-880
93. Tran H, Gourrier N, Lemercier-Neuillet C, Dhaenens CM, Vautrin A, Fernandez-Gomez FJ. et al. Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J Biol Chem. 2011;286(18):16435-16446
94. Chen YS, Liu CW, Lin YC, Tsai CY, Yang CH, Lin JC. The SRSF3-MBNL1-Acin1 circuit constitutes an emerging axis to lessen DNA fragmentation in colorectal cancer via an alternative splicing mechanism. Neoplasia. 2020;22(12):702-713
95. Ke H, Zhao L, Zhang H, Feng X, Xu H, Hao J. et al. Loss of TDP43 inhibits progression of triple-negative breast cancer in coordination with SRSF3. Proc Natl Acad Sci U S A. 2018;115(15):E3426-E3435
96. Cui M, Allen MA, Larsen A, Macmorris M, Han M, Blumenthal T. Genes involved in pre-mRNA 3'-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc Natl Acad Sci U S A. 2008;105(43):16665-16670
97. Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279(34):35788-35797
98. Chen W, Jia Q, Song Y, Fu H, Wei G, Ni T. Alternative Polyadenylation: Methods, Findings, and Impacts. Genomics Proteomics Bioinformatics. 2017;15(5):287-300
99. Shen T, Li H, Song Y, Li L, Lin J, Wei G. et al. Alternative polyadenylation dependent function of splicing factor SRSF3 contributes to cellular senescence. Aging (Albany NY). 2019;11(5):1356-1388
100. Schwich OD, Blumel N, Keller M, Wegener M, Setty ST, Brunstein ME. et al. SRSF3 and SRSF7 modulate 3'UTR length through suppression or activation of proximal polyadenylation sites and regulation of CFIm levels. Genome Biol. 2021;22(1):82
101. Hardy JG, Norbury CJ. Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation. Biochem Soc Trans. 2016;44(4):1051-1057
102. Park SK, Jeong S. SRSF3 represses the expression of PDCD4 protein by coordinated regulation of alternative splicing, export and translation. Biochem Biophys Res Commun. 2016;470(2):431-438
103. Ratnadiwakara M, Archer SK, Dent CI, Ruiz De Los Mozos I, Beilharz TH, Knaupp AS. et al. SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife. 2018;7:e37419
104. Mure F, Corbin A, Benbahouche NEH, Bertrand E, Manet E, Gruffat H. The splicing factor SRSF3 is functionally connected to the nuclear RNA exosome for intronless mRNA decay. Sci Rep. 2018;8(1):12901
105. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507-519
106. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14(5):e1007412
107. Wang ZW, Pan JJ, Hu JF, Zhang JQ, Huang L, Huang Y. et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep. 2022;39(6):110813
108. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311
109. Fernandez N, Cordiner RA, Young RS, Hug N, Macias S, Caceres JF. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114
110. Wang C, Li L, Yin Z, Zhang Q, Zhao H, Tao R. et al. An indel polymorphism within pre-miR3131 confers risk for hepatocellular carcinoma. Carcinogenesis. 2017;38(2):168-176
111. Kim HR, Hwang SJ, Shin CH, Choi KH, Ohn T, Kim HH. SRSF3-regulated miR-132/212 controls cell migration and invasion by targeting YAP1. Exp Cell Res. 2017;358(2):161-170
112. Kim HR, Shin CH, Lee H, Choi KH, Nam DH, Ohn T. et al. MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3. Oncotarget. 2017;8(5):8342-8355
113. DeLigio JT, Stevens SC, Nazario-Munoz GS, MacKnight HP, Doe KK, Chalfant CE. et al. Serine/Arginine-Rich Splicing Factor 3 Modulates the Alternative Splicing of Cytoplasmic Polyadenylation Element Binding Protein 2. Mol Cancer Res. 2019;17(9):1920-1930
114. Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N. et al. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2014;33(11):1407-1417
115. Kim YJ, Kim BR, Ryu JS, Lee GO, Kim HR, Choi KH. et al. HNRNPA1, a Splicing Regulator, Is an Effective Target Protein for Cervical Cancer Detection: Comparison With Conventional Tumor Markers. Int J Gynecol Cancer. 2017;27(2):326-331
116. Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K. et al. SRSF3, a Splicer of the PKM Gene, Regulates Cell Growth and Maintenance of Cancer-Specific Energy Metabolism in Colon Cancer Cells. Int J Mol Sci. 2018;19(10):3012
117. Peiqi L, Zhaozhong G, Yaotian Y, Jun J, Jihua G, Rong J. Expression of SRSF3 is Correlated with Carcinogenesis and Progression of Oral Squamous Cell Carcinoma. Int J Med Sci. 2016;13(7):533-539
118. He X, Ee PL, Coon JS, Beck WT. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res. 2004;10(14):4652-4660
119. He X, Arslan AD, Pool MD, Ho TT, Darcy KM, Coon JS. et al. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2011;30(3):356-365
120. Wang H, Lekbaby B, Fares N, Augustin J, Attout T, Schnuriger A. et al. Alteration of splicing factors' expression during liver disease progression: impact on hepatocellular carcinoma outcome. Hepatol Int. 2019;13(4):454-467
121. Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, Dennewitz MB. et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276(1):74-88
122. Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet. 2009;18(19):3696-3707
123. Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF. et al. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32(34):3933-3943
124. Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH. et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 2016;126(1):68-84
125. Kim J, Park RY, Chen JK, Kim J, Jeong S, Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5'-UTR region. Cell Death Differ. 2014;21(3):481-490
126. Jang HN, Lee M, Loh TJ, Choi SW, Oh HK, Moon H. et al. Exon 9 skipping of apoptotic caspase-2 pre-mRNA is promoted by SRSF3 through interaction with exon 8. Biochim Biophys Acta. 2014;1839(1):25-32
127. Lee BP, Pilling LC, Emond F, Flurkey K, Harrison DE, Yuan R. et al. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell. 2016;15(5):903-913
128. Turnquist C, Horikawa I, Foran E, Major EO, Vojtesek B, Lane DP. et al. p53 isoforms regulate astrocyte-mediated neuroprotection and neurodegeneration. Cell Death Differ. 2016;23(9):1515-1528
129. Zeng S, Shen WH, Liu L. Senescence and Cancer. Cancer Transl Med. 2018;4(3):70-74
130. Zhou L, Guo J, Jia R. Oncogene SRSF3 suppresses autophagy via inhibiting BECN1 expression. Biochem Biophys Res Commun. 2019;509(4):966-972
131. He X, Zhang P. Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol Cancer. 2015;14:158
132. Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5(2):357-366
133. Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261(29):13807-13812
134. Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230-233
135. Wang Z, Chatterjee D, Jeon HY, Akerman M, Vander Heiden MG, Cantley LC. et al. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J Mol Cell Biol. 2012;4(2):79-87
136. Jiang H, Zhang S, Song T, Guan X, Zhang R, Chen X. Trichostatin a Protects Dendritic Cells Against Oxygen-Glucose Deprivation via the SRSF3/PKM2/Glycolytic Pathway. Front Pharmacol. 2018;9:612
137. Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34(9):518-530
138. Buoso E, Ronfani M, Galasso M, Ventura D, Corsini E, Racchi M. Cortisol-induced SRSF3 expression promotes GR splicing, RACK1 expression and breast cancer cells migration. Pharmacol Res. 2019;143:17-26
139. Buoso E, Galasso M, Ronfani M, Serafini MM, Lanni C, Corsini E. et al. Role of spliceosome proteins in the regulation of glucocorticoid receptor isoforms by cortisol and dehydroepiandrosterone. Pharmacol Res. 2017;120:180-187
140. Yan XB, Tang CH, Huang Y, Fang H, Yu ZQ, Wu LM. et al. Alternative splicing in exon 9 of glucocorticoid receptor pre-mRNA is regulated by SRp40. Mol Biol Rep. 2010;37(3):1427-1433
141. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871-884
142. Yoon SP, Kim HH, Kim J, Park RY, Ohn T. Regulation of cellular RNA nano-particle assembly by splicing factor SRp20. J Nanosci Nanotechnol. 2013;13(1):184-187
143. Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18(24):3574-3582
144. Gao X, Gao C, Liu G, Hu J. MAP4K4: an emerging therapeutic target in cancer. Cell Biosci. 2016;6:56
145. Hassn Mesrati M, Syafruddin SE, Mohtar MA, Syahir A. CD44: A Multifunctional Mediator of Cancer Progression. Biomolecules. 2021 11(12)
146. Zhu S, Chen Z, Katsha A, Hong J, Belkhiri A, El-Rifai W. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene. 2016;35(14):1847-1856
147. Guo L, Ke H, Zhang H, Zou L, Yang Q, Lu X. et al. TDP43 promotes stemness of breast cancer stem cells through CD44 variant splicing isoforms. Cell Death Dis. 2022;13(5):428
148. Koltai T, Reshkin SJ, Carvalho TMA, Di Molfetta D, Greco MR, Alfarouk KO. et al. Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma: A Physiopathologic and Pharmacologic Review. Cancers (Basel). 2022;14(10):2486
149. Chen Y, Yang M, Meng F, Zhang Y, Wang M, Guo X. et al. SRSF3 Promotes Angiogenesis in Colorectal Cancer by Splicing SRF. Front Oncol. 2022;12:810610
150. Moura-Alves P, Neves-Costa A, Raquel H, Pacheco TR, D'Almeida B, Rodrigues R. et al. An shRNA-based screen of splicing regulators identifies SFRS3 as a negative regulator of IL-1beta secretion. PLoS One. 2011;6(5):e19829
151. Boutej H, Rahimian R, Thammisetty SS, Beland LC, Lalancette-Hebert M, Kriz J. Diverging mRNA and Protein Networks in Activated Microglia Reveal SRSF3 Suppresses Translation of Highly Upregulated Innate Immune Transcripts. Cell Rep. 2017;21(11):3220-3233
152. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677-704
153. Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J. et al. TGFbeta1-Mediated SMAD3 Enhances PD-1 Expression on Antigen-Specific T Cells in Cancer. Cancer Discov. 2016;6(12):1366-1381
154. Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G. et al. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov. 2015;5(12):1282-1295
155. Ben C, Wu X, Takahashi-Kanemitsu A, Knight CT, Hayashi T, Hatakeyama M. Alternative splicing reverses the cell-intrinsic and cell-extrinsic pro-oncogenic potentials of YAP1. J Biol Chem. 2020;295(41):13965-13980
156. Majerciak V, Zheng ZM. KSHV ORF57, a protein of many faces. Viruses. 2015;7(2):604-633
157. Majerciak V, Lu M, Li X, Zheng ZM. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA. 2014;20(11):1747-1758
158. Fitzgerald KD, Semler BL. Re-localization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathog. 2011;7(7):e1002127
159. Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol. 2002;76(23):12008-12022
160. Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71(8):6243-6246
161. Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26(2):459-467
162. Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401(2):155-164
163. Verma D, Bais S, Gaillard M, Swaminathan S. Epstein-Barr Virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J Virol. 2010;84(22):11781-11789
164. Dumont AA, Dumont L, Zhou D, Giguere H, Pileggi C, Harper ME. et al. Cardiomyocyte-specific Srsf3 deletion reveals a mitochondrial regulatory role. FASEB J. 2021;35(5):e21544
165. Wong J, Garner B, Halliday GM, Kwok JB. Srp20 regulates TrkB pre-mRNA splicing to generate TrkB-Shc transcripts with implications for Alzheimer's disease. J Neurochem. 2012;123(1):159-171
166. Heazlewood S, Ahmad T, Mohenska M, Guo BB, Gangatirkar P, Josefsson EC. et al. RNA Binding Protein SRSF3 confers an essential role in megakaryocyte maturation and platelet production. Blood. 2021;139(9):1359-1373
167. Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336
168. Tao H, Szeszel-Fedorowicz W, Amir-Ahmady B, Gibson MA, Stabile LP, Salati LM. Inhibition of the splicing of glucose-6-phosphate dehydrogenase precursor mRNA by polyunsaturated fatty acids. J Biol Chem. 2002;277(34):31270-31278
169. Griffith BN, Walsh CM, Szeszel-Fedorowicz W, Timperman AT, Salati LM. Identification of hnRNPs K, L and A2/B1 as candidate proteins involved in the nutritional regulation of mRNA splicing. Biochim Biophys Acta. 2006;1759(11-12):552-561
170. Szeszel-Fedorowicz W, Talukdar I, Griffith BN, Walsh CM, Salati LM. An exonic splicing silencer is involved in the regulated splicing of glucose 6-phosphate dehydrogenase mRNA. J Biol Chem. 2006;281(45):34146-34158
171. Cyphert TJ, Suchanek AL, Griffith BN, Salati LM. Starvation actively inhibits splicing of glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS element bound by hnRNP K. Biochim Biophys Acta. 2013;1829(9):905-915
172. Guo J, Che X, Wang X, Jia R. Inhibition of the expression of oncogene SRSF3 by blocking an exonic splicing suppressor with antisense oligonucleotides. RSC Advances. 2018;8:7159-7163
173. Sun Y, Yan L, Guo J, Shao J, Jia R. Downregulation of SRSF3 by antisense oligonucleotides sensitizes oral squamous cell carcinoma and breast cancer cells to paclitaxel treatment. Cancer Chemother Pharmacol. 2019;84(5):1133-1143
174. Zhang Y, Wang M, Meng F, Yang M, Chen Y, Guo X. et al. A novel SRSF3 inhibitor, SFI003, exerts anticancer activity against colorectal cancer by modulating the SRSF3/DHCR24/ROS axis. Cell Death Discov. 2022;8(1):238
175. Chang JG, Yang DM, Chang WH, Chow LP, Chan WL, Lin HH. et al. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS One. 2011;6(6):e18643
176. Anderson ES, Lin CH, Xiao X, Stoilov P, Burge CB, Black DL. The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. RNA. 2012;18(5):1041-1049
177. Avanes A, Lenz G, Momand J. Darpp-32 and t-Darpp protein products of PPP1R1B: Old dogs with new tricks. Biochem Pharmacol. 2019;160:71-79
178. Baek JY, Yun HH, Jung SY, Lee J, Yoo K, Lee JH. SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA. Cells. 2020;9(10):2325
Author contact
![]() Corresponding authors: jiarongedu.cn; zhengtnih.gov.
Corresponding authors: jiarongedu.cn; zhengtnih.gov.

 Global reach, higher impact
Global reach, higher impact