10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(13):5293-5311. doi:10.7150/ijbs.96469 This issue Cite
Review
Three-Dimensional In Vitro Cell Cultures as a Feasible and Promising Alternative to Two-Dimensional and Animal Models in Cancer Research
1. Laboratory of Molecular Pathology, Department of Health Sciences, Università del Piemonte Orientale, Via Solaroli 17, 28100, Novara, Italy.
2. Stephenson Cancer Center, The University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA.
Received 2024-3-20; Accepted 2024-8-25; Published 2024-9-30
Abstract
Cancer represents one of the diseases with the highest mortality rate worldwide. The burden of cancer continues to increase, not only affecting the health-related quality of life of patients but also causing an elevated global financial impact. The complexity and heterogeneity of cancer pose significant challenges in research and clinical practice, contributing to increase the failure rate of clinical trials for antitumoral drugs. This is partially due to the fact that preclinical models still present important limitations in faithfully recapitulating human tumors to serve as reliable indicators of drug effectiveness. Up to now, research and development strategies employ expensive animal models (including the so-called “humanized mice”) that not only raise ethical concerns, but also frequently fail to accurately predict responses to anticancer drugs because they do not faithfully replicate human physiology as well as the patient's tumor microenvironment. On the other side, traditional two-dimensional (2D) cell cultures fail to adequately reproduce the structural organization of tumor and the cellular heterogeneity found in vivo. The growing necessity to develop more accurate cancer models has increasingly emphasized the importance of three-dimensional (3D) in vitro cell cultures, such as cancer-derived spheroids and organoids, as promising alternatives to bridge the gap between 2D and animal models. In this review, we provide a brief overview focusing on 3D in vitro cell cultures as preclinical models capable of properly reproducing the tissue organization, biological composition, and complexity of in vivo tumors in a fine-tuned microenvironment. Despite their limitations, these models collectively enhance our understanding of the mechanisms underlying cancer and may offer the potential for a more reliable assessment of drug efficacy before clinical testing and, consequently, improve therapeutic outcomes for cancer patients.
Keywords: tumor spheroids, organoids, microfluidics, organ-on-a-chip, 3D models, humanized mice
Introduction
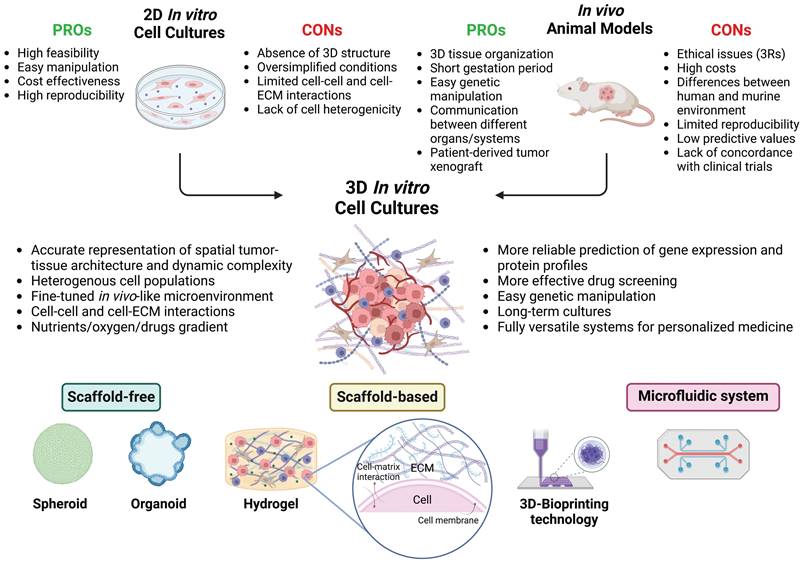
Despite the development of improved prevention and early detection methods, as well as numerous advances in therapeutic strategies over the last decades, cancer remains one of the leading causes of death worldwide.1 Cancer is a multifactorial disease characterized by high complexity and heterogeneity, which makes experimental reproduction difficult and involves continuous therapeutic adaptations in the clinic.2,3 Due to cancer heterogeneity, a “one drug fits all” therapeutic approach is not feasible, as evidenced by the fact that current standard treatments are effective only in a subset of the patients' population. Tumors can exhibit intrinsic differences in the genetic background and may express different proteins in one patient compared to another. This variability highlights the urgent need for precision and personalized medicine.4 To this aim, the production of highly efficient and cost-effective drugs emerges as a crucial point in the future development of therapeutic approaches. Drug discovery is an arduous, meticulous, and expensive process; it is estimated that the cost of innovating and developing a new drug in the United States alone is more than $600 million, with an average timeline of 10-15 years.5 Therefore, there is a significant, unmet need to develop inexpensive and reliable preclinical platforms to accelerate the anticancer drug discovery pipeline to improve the outcome of cancer patients.6
Recapitulative disease models serve as important experimental tools, particularly in cancer research, where new drugs exhibit higher failure rates in clinical trials compared to other fields. In fact, only 5% of new anticancer compounds receive clinical approval, with the majority failing due to issues related to toxicity and insufficient effectiveness. This limited clinical efficacy can be partly attributed to the absence of preclinical models that closely reproduce tumor architecture, pathophysiology, and the crosstalk between cancer cells and the surrounding microenvironment. This represents the major shortcoming of the correlation between preclinical in vitro and in vivo results and the data obtained during the clinical trials.7,8 Therefore, a potential approach to mitigate the high attrition of anticancer drugs is to employ preclinical models that more closely represent in vivo human tumors, thereby obtaining more precise indications in terms of drug efficiency.9,10 To date, various models and technical approaches have been used to investigate and study the mechanisms underlying the key hallmarks of cancer.2,11
The two-dimensional (2D) in vitro cell cultures are traditional systems where cells grow as an adherent monolayer on a solid and flat plastic surface. These models have played a crucial role in setting the current knowledge of cancer biology research and continue to be widely used due to their high availability, easy manipulation, elevated level of reproducibility, and cost-effectiveness.3,12 However, 2D cancer cell cultures represent reductive models where cells are cultured under oversimplified and unrealistic conditions, thus failing to accurately mimic the real physiology of an in vivo tissue. They cannot reproduce the complex three-dimensional (3D) structure of a tumor or the dynamic interactions between cancer cells and the tumor microenvironment (TME). Moreover, cancer cells cultured as a monolayer on a flat plastic surface display significant alterations in crucial cellular signaling pathways and changes in their responses to stimuli. The 2D models then present further limitations: i) they do not preserve the real shape and polarization of cancer cells, ii) there is a marked lack of tumor cell heterogeneity, iii) they provide equal and unrestricted accessibility to essential nutrients, oxygen, growth factors, and metabolites, and iv) they offer an altered representation of critical cellular activities such as proliferation, migration, invasion, differentiation, gene and protein expression, response to stimuli and drug sensitivity. Furthermore, to maintain regular cell growth and ensure the supply of essential nutrients, adherent cells must undergo periodic trypsinization. This continuous procedure could lead to different and unpredictable genetic and epigenetic changes over time, consequently impacting cells' phenotype and behavior, such as growth and their response to both external and internal stimuli.8,12-15 Another aspect that limits the reproducibility of data in cultured cancer cultures is that the same cell line from different laboratories presents differences in its genetic and epigenetic background as a consequence of the long-term culture under different cultivation conditions, which ultimately leads to clonal evolution and selection of different subclones.16
Another widely employed tool in cancer research is represented by animal models, which act as an important link between in vitro studies and clinical experimentation.17 These models enable a deeper understanding of the complex nature of cancer biology and the implementation of novel strategies in terms of prevention, diagnosis, and treatment.18 Unlike 2D cell cultures, they effectively recapitulate the tissue structural organization and provide a system-level analysis.19 Specifically, mouse models represent the most commonly utilized in vivo systems due to their low cost, short gestation period, ease of genetic manipulation and the possibility to grow cancer cells from patients, generating patient-derived tumor xenografts (PDTXs). PDTXs can recreate the histological and molecular features, as well as the inter/intratumor heterogeneity of a patient's tumor, thus allowing the investigation of anticancer drug effectiveness. However, these models present several limitations ascribed to their dependence on immunodeficient hosts and the expensive costs related to their maintenance, long-term engraftment process, and molecular profiling. In addition, certain biological shortcomings hinder the efficacy and predictive value of murine models, as well as other types of in vivo studies. Genomic and pathophysiological differences between animals and humans allow the study of cancer hallmarks under conditions similar, but not identical, to those of patients. For instance, in the case of PDTXs, tumor engraftment might not occur, or metastatic patterns might not be faithfully replicated as in humans. Furthermore, the murine and human TMEs significantly differ, and mice also tend to substitute the exogenous tumor stroma and immune cells with their own. To overcome these limitations, humanized mice models consisting in severely immunodeficient mice (usually NOD-SCID) in which both patient's derived tumor cells and immune cells are co-engrafted could be employed. 20,21 One important drawback of these models is the lack of an adequate innate and MHC-dependent immune response and the inability to reconstruct a TME with a broad range of cytokines and stromal cells that can faithfully resemble that in the native patient's derived tumor. 21,22 This has relevance in terms of cancer stem cell renewal, angiogenesis, metastasization and tumor dormancy, all features that influence the natural growth and progression of the tumor and that impact the response to the therapy, thus affecting the translational value of immune-oncological research and drug testing in such animals. 21 Additionally, factors such as the gender, the number of animals enrolled in the study, their age, and the level of stress to which they are exposed can vary between laboratories and may significantly influence the outcome of the experiments.8,19,23 Considering these factors, the evaluation of drug effectiveness and toxicity in preclinical animal testing is not a foolproof indicator of its effects when translated to human clinical trials and raises ethical concerns as well.24 It is a fact that despite the preclinical test with these rodent models, more than 90% of drugs are revealed to be toxic to patients. 25
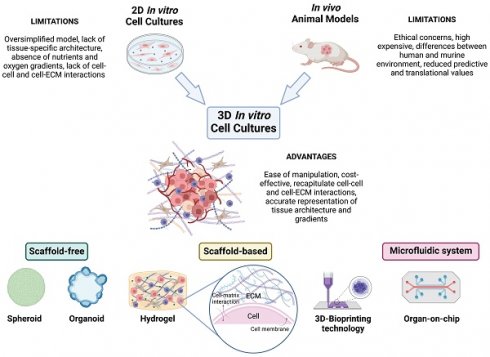
The application of 3Rs' principle (e.g., Replacement, Reduction, Refinement) in research studies promotes a decrease or elimination in the use of laboratory animals, suggesting the need to develop and use new and more innovative models to overcome these limitations.8,15,26 In agreement, under recent legislation, the FDA (United States Food and Drug Administration) no longer requires animal testing to approve new drugs. Over the past decade, a pioneering approach to modeling cancer that incorporates the most recent advances in the fields of cancer research, pharmacology, tissue engineering, biomaterials, and nanotechnologies has progressively emerged. 3D in vitro cancer models are preclinical models that can accurately replicate the tumor-tissue architecture, biological composition, and dynamic complexity of in vivo tumors in a fine-tuned microenvironment. This facilitates a deeper comprehension of the mechanisms underlying tumor aetiology, growth, invasion, and metastasis, as well as the evaluation of drug efficacy before clinical testing.27 The growing need for more accurate cancer models highlights the relevance of these 3D cell culture techniques as a promising alternative for bridging the gap between 2D cell cultures and animal models (Figure 1). In fact, several 3D culture models have been developed and employed over the years.28,29
The focus of the present review is on the two widely employed 3D models in cancer research, namely spheroids and organoids, excluding the description of the methods by which they can be generated.
Tumor Spheroids
Spheroids, first described by Sutherland and colleagues in the early 1970s, represent one of the most consolidated 3D culture methods for studying cancer biology. They are closely packed clusters of cells that faithfully reproduce several important properties of in vivo tumors, including structural organization, cellular heterogeneity, cell signaling pathways, deposition of the extracellular matrix (ECM), cell-cell and cell-ECM communications, growth kinetics, genomic and proteomic expression profiles, and pharmacological resistance.30,31
Spherical cancer models can be classified into different types based on cellular origin and methods of preparation, as widely described in the study published by Weiswald and colleagues.32 Among the reported models, multicellular tumor spheroids (MCTSs) emerge as the most extensively characterized and widely employed for recapitulating a variety of solid tumors, including breast, cervical, colon, lung, pancreatic, prostate, and adrenal cancers 33-39, among many others. MCTSs are self-assembling cellular aggregates consisting of primary cells or cell lines and mimic the physiological architecture of the tumor mass in vivo with its metabolic and proliferative gradients, acting as a clinically significant model for drug resistance studies. Moreover, the cell clonality, ease of maintenance, and simplicity of genetic manipulation make the 3D model the most suitable approach for high-throughput anticancer drug screening.40-42
MCTSs formation can be easily obtained in non-adherent conditions (like ultra-low attachment plates, continuous agitation and/or centrifugation) or in the presence of an exogenous scaffold.43 Depending on ECM composition and cadherin types and concentration (which vary for different cell types), spheroids assembly requires cell-cell adhesion and/or cell-ECM interactions that are orchestrated from homophilic cadherin-cadherin binding, integrins-ECM interactions, and cytoskeletal proteins remodeling. 44 In scaffold-based approaches, cells can proliferate dispersed in, or they can adhere to, ECM-mimicking acellular hydrogels.45,46 The scaffold can influence the mechanical and biochemical signals, facilitates cell-cell and cell-ECM interactions, and mimics the hypoxic and nutrient deprivation conditions of the native TME. 47 These models are particularly suitable for those tumors characterized by abundant ECM deposition and strictly dependent on it for their growth in vivo, like breast 48, pancreatic 49, lung 50 and liver cancers 51, for culturing primary patient-derived cancer cells, and for drug screening purposes. 52,53 In contrast, scaffold-free techniques mainly consist of culturing the cells under conditions that promote strong cell-to-cell interactions, facilitating cancer cell aggregation and ECM deposition. This method represents an excellent in vitro system for studies on tumor-specific processes like angiogenesis, invasion and metastasis. 54 Additionally, scaffold-free approach is a suitable platform to model the transcoelomic growth of peritoneal tumors (e.g., ovarian and colorectal cancers). 54-57
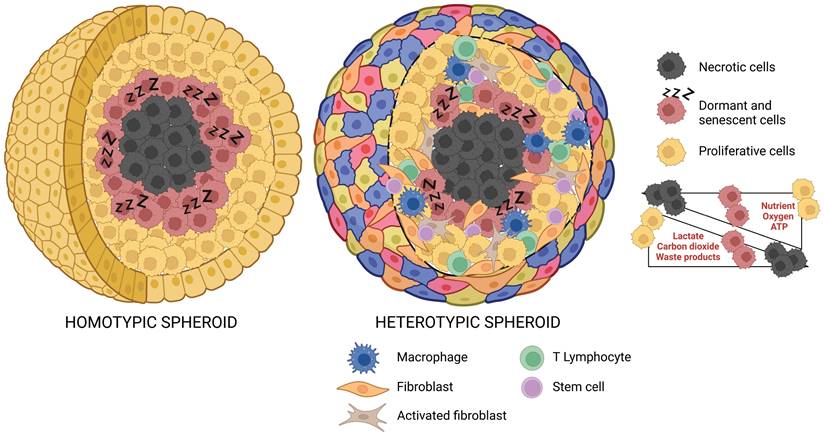
Tumor spheroids can be distinguished as homotypic, when composed exclusively of tumor cells, or heterotypic, when tumor cells are co-cultured with stromal cells, such as fibroblasts, endothelial cells, and immune cells.31,58 Remarkably, heterotypic spheroids allow to dissect the dynamic interactions between cancer cells and the cellular components of the TME, which play a crucial role in cancer metabolism and response to therapy. For instance, a heterotypic spheroid model (combined of pheochromocytoma cells and primary cancer-associated fibroblasts (CAFs)) was used to test the differential response of wild-type and SDHB/SDHD knock-down pheochromocytoma cells to the pro-migratory factors released by CAFs.59 To be noted, the 3D heterotypic spheroids displayed increased tumorigenic potential in terms of migratory capabilities compared to that of 3D homotypic cancer spheroids. 60 Giustarini and colleagues used 3D heterotypic tumor spheroids made of pancreatic ductal adenocarcinoma (PDAC) cells, endothelial cells, pancreatic stellate cells (PSC), and monocytes, which resembled some critical features of patients' PDAC immune microenvironment (e.g., immunosuppressive phenotype), for testing chemo- and immuno-therapeutics. 37
3D In Vitro Models: Bridging 2D Cell Cultures and Animal Models. This image illustrates how 3D in vitro models bridge the gap between traditional 2D cell cultures and in vivo animal models. Unlike 2D cultures, which lack three-dimensional interactions and spatial organization, 3D cellular systems more accurately replicate the complex architecture and properties of tumor tissues. This realistic microenvironment enables precise studies of tumor biology and drug responses. In contrast to in vivo models, which may be limited by species-specific differences, 3D models can be tailored to closely mimic human tissues, enhancing their translational relevance and predictive accuracy. Ethically, 3D models offer advantages by reducing reliance on animal testing, adhering to the principles of the 3Rs (Replacement, Reduction, Refinement), and providing a more humane alternative for research. Various approaches can be used to develop 3D tumor models, including scaffold-free, scaffold-based, 3D-bioprinting, and microfluidic methods (created with Biorender).
The employment of microfluidic systems or 3D-bioprinting technologies enable the development of even more sophisticated 3D cancer models, generating systems with complex architecture, physiological microenvironmental conditions, vasculature-like perfusion, precise control over chemical gradient flows, and mechanical forces.15,40,61 These techniques are also applicable for establishing organoids.
Tumor spheroids with a diameter exceeding 500 µm accurately reproduce avascular tumors or micrometastases, displaying a stratified structure with cells having various phenotypic, functional, and metabolic behaviors. Specifically, these spheroids present a well-organized spatial architecture that comprises an outer proliferative layer, an intermediate zone composed of quiescent and senescent cells, and an inner apoptotic and necrotic core resulting from the altered distribution of nutrients and oxygen (Figure 2). Typically, the diffusion gradient is confined to a range of 150-200 µm. 62 Indeed, the progression towards the spheroid core leads to a reduction of oxygen, nutrients, and pH levels, together with an increase of carbon dioxide, lactate, and waste products.2,3,63,64 Tumor cells can adapt to hypoxic conditions, by shifting from oxidative phosphorylation to anaerobic glycolysis and converting pyruvate into lactate to obtain energy. As a result, the release of lactate increases the acidification of the spheroid's inner regions.65,66
The structural organization and physiological characteristics of tumor spheroids strongly influence the response to therapy, the cell signaling pathways, and the profiles of gene and protein expression.31 Recently, our group demonstrated that the expression of the lysosomal protease cathepsin D (CD) is differentially modulated between 2D and 3D cell culturing, and this reflects on the survival efficiency of neuroblastoma (NB) cells. 67 Interestingly, CD-overexpressing NB cells were favored to grow in suspension (3D), while the CD knocked-down cells were favored for the growth in 2D, and when cells were switched from 2D to 3D and back to 2D culture conditions the surviving clones adjusted the expression of CD accordingly. 67 This example highlights how the culture condition influences the gene and protein expression.
Tumor spheroids can be exploited also to study in vitro the phenotypical changes and cancer cell behavior in relation to microbiota/host interactions. For instance, 3D colorectal cancer spheroids allowed to study how pro-inflammatory cytokines and probiotics impacted on cell proliferation and migration. 68,69
The hypoxic environment within tumor spheroids can induce chemo-radio resistance through various mechanisms. Firstly, hypoxic conditions cause an increase in hypoxia-inducible factors (HIFs) levels. Consistently, the expression of HIF-1α protein was detected in HeLa spheroids but not in the 2D culture counterpart.70 HIFs can induce the upregulation of the multidrug resistance (MDR1) gene, which encodes P-glycoprotein (P-gp), a cell membrane protein that actively pumps drugs or other molecules outside the cell. One study observed that the expression of HIF-1 in MCF-7 breast cancer spheroids caused an upregulation of P-gp, which, in turn, reduced the accumulation of doxorubicin within the spheroids; in contrast, no significant changes in HIF-1 and P-gp expression were observed in 2D-cultured cells.71 Furthermore, HIF-1 can regulate the expression of the vascular endothelial growth factor (VEGF), which is highly expressed within the hypoxic regions of spheroids where it contributes to drug resistance mechanisms.72,73 A375 melanoma spheroids were shown to exhibit higher expression of HIF-1 and VEGF compared to the same cells cultured as monolayers, and this variation affected the sensitivity of the cells to vemurafenib.73 It is known that hypoxia can impair the effectiveness of radiotherapy because DNA lesions, derived from reactive oxygen species (ROS) generated during water radiolysis, react with oxygen to generate stable DNA peroxides. This phenomenon contributes to pronounced radio resistance in tumor cells located in the inner part of the spheroid.74,75 Lastly, the hypoxic conditions within the spheroid are deleterious for drugs that cause cell membrane and DNA damage through the generation of ROS, such as doxorubicin 38,76,77 and cisplatin. 78 Moreover, drugs targeting highly proliferative cells (including carboplatin, cisplatin, doxorubicin, oxaliplatin, methotrexate, and paclitaxel) present limited efficacy against senescent and necrotic cells located in the inner regions of the spheroids.79 For instance, compared to 2D cell cultures, dense spheroids of BT-549, BT-474, and T-47D breast cancer cells showed increased resistance to doxorubicin and paclitaxel associated with elevated levels of hypoxia, high proportion of G0 dormant cells, and reduced expression of PARP and caspase-3. 33 In addition, low pH affects the efficacy of several anticancer compounds, such as doxorubicin, vinblastine, methotrexate, and anthraquinone, by impairing intracellular uptake.80-82
Finally, the strong E-cadherin-driven interactions between cancer cells and the secretion of ECM proteins (collagen, fibronectin, laminin, elastin, tenascin) determine an increase of spheroid density, forming a physical barrier that hampers the transport of therapeutic agents into the spheroid mass. Simultaneously, the elevated interstitial fluid pressure hinders the intake and distribution of antitumoral compounds.10,83,84 Several studies reviewed by Nunes et al. have shown that E-cadherin expression is more pronounced in spheroid cultures than in 2D cell cultures.10 Additionally, to confirm the role of E-cadherin on drug resistance in spheroids, an anti-E-cadherin monoclonal antibody (SHE78-7) was utilized to inhibit its function. The study shows that the administration of SHE78-7 increases the intracellular accumulation of chemotherapeutics (5-fluorouracil, etoposide, paclitaxel, and vinblastine) in HT-29 human colorectal cancer spheroids.85
The different gene and protein expression between 2D and 3D models provides an explanation for the distinct behaviors observed in 3D-cultured cells regarding growth, proliferation, migration, invasion, and drug sensitivity, when compared to cells grown in 2D.46 For instance, in a monolayer, several genes that promote growth and proliferation are frequently upregulated in comparison with their corresponding tissue origins, while genes that restrain these phenotypes tend to be suppressed. Therefore, cells grown in 2D cultures generally display a higher rate of proliferation in contrast to cells cultivated in 3D models. Although cells lose many of their original features when extracted from the primary tumor and cultured in a 2D system, the reintroduction of these cells into an in vivo-like environment largely restores the original features in terms of morphology, proliferation, and gene/protein expression.86,87 Differences in gene and protein expression profiles between 2D and 3D cultures have been reported for various types of cancers, including melanoma 88, colorectal cancer 89, mesothelioma 90, liver hepatocellular carcinoma 91 , and neuroblastoma 67. Ghosh and colleagues performed a comparative analysis of the expression patterns of 179 genes responsible for encoding chemokines, pro-angiogenic factors, and cell-adhesion molecules in both 2D and 3D models of melanoma cells. They detected significant upregulation of several genes that play important roles in promoting the progression, invasion, and metastasis of skin cancer in the 3D spheroids.88
miRNAs, which are key regulators of gene expression, can also be modulated in 3D models.92 Extracellular vesicles (EVs) released from 3D spheroids display differences in terms of secretion dynamics and molecular components (RNA and DNA) compared with EVs derived from 2D monolayers.93 In this context, miRNA expression profiles of EVs derived from 3D cultures of HeLa cervical cancer cells closely resembled those of circulating EVs isolated from the plasma of cervical cancer patients, with a high similarity of about 96%, in contrast to the miRNA expression patterns of EVs obtained from 2D cultured HeLa cells. Furthermore, culture and growth conditions had no impact on the genomic information carried by EVs, as demonstrated by DNA sequencing analysis.93 In addition, 3D models were useful for investigating miRNA-mediated regulatory mechanisms in ovarian cancer. Yoshimura and colleagues studied the effect of miR-99a-5p, a microRNA overexpressed in epithelial ovarian carcinoma (EOC), on peritoneal dissemination. They utilized human peritoneal mesothelial cells (HPMCs) treated with EOC-derived exosomes. Results revealed that upregulation of miR-99a-5p in HPMCs promoted EOC invasion by inducing upregulation of fibronectin and vitronectin, suggesting its potential utility as an EOC biomarker in serum and as a potential therapeutic target.94 3D ovarian cancer spheroids, mimicking peritoneal metastases dissemination, have been employed in our laboratories to study autophagy-dependent cancer cell dormancy and how this was interrupted by IL-6-mediated upregulation of the oncomiRNA miR-1305.95 Furthermore, the expression of miRNAs in 3D cultures compared to 2D cultures has been investigated in different breast cancer cell lines. Nguyen and colleagues examined the miRNA expression profile in 3D cultures versus 2D cultures in the breast cancer cell lines MCF-7 (non-invasive) and MDA-MB-231 (invasive). They found that 49 miRNAs exhibited differential expression in the MCF-7 cell line when cultured in 3D compared to 2D, while in the MDA-MB-231 cell line 28 miRNAs displayed differential levels.96
Finally, differences in crucial cellular signaling pathways may exist between 2D- and 3D-cultured cells. Compared to their 2D counterparts, colon cancer cell lines cultured in a 3D environment exhibit a reduced activation of the AKT/mTOR/S6K pathway, which plays an important role in carcinogenesis, cancer cell migration, and resistance to therapies.97 Similarly, 3D cultures of ER+/Her2+ breast cancer cells were less responsive to hormonal and anti-HER2 treatments compared to 2D cultures due to a shift from the AKT to the MAPK pathway.98
Recently we have proposed a variant of heterotypic/homotypic spheroids made of a mixture of subclones of cancer cells genetically engineered to express different levels of a protein as an in vitro model that could bona fide represent the genetic changes occurring during clonal evolution. 67 The model allowed to determine which clone, among the ones hyper-expressing or silenced for cathepsin D, would overtake the other depending on the culture condition.
Spatial Organization of Homotypic and Heterotypic Tumor Spheroids. This schematic representation illustrates the 3D architecture of both homotypic and heterotypic tumor spheroids. The homotypic spheroid is comprised of a single cancer cell type, while the heterotypic spheroid includes multiple cell types, reflecting the complex nature of the tumor microenvironment. Both spheroids exhibit distinct regions: the outer proliferative zone, the intermediate quiescent (dormant) zone, and the central necrotic core. The figure also highlights the gradient distribution of essential factors such as nutrients, oxygen, lactate, carbon dioxide, and waste products across these zones, providing insights into the varying conditions within different regions of the spheroid (created with BioRender).
Tumor Organoids
Organoids are miniaturized replicas of tissues or organs, originating from stem cells with the ability to differentiate and self-assemble into in vitro 3D structures, faithfully mimicking the morphology and functionality of their in vivo counterparts.99-100 Organoids can be derived from various types of stem cells, including induced pluripotent stem cells (iPSCs), adult stem cells (ASCs), or embryonic stem cells (ESCs).14,101 Like spheroids, organoids are cultured in the presence of an ECM-mimicking scaffold that provides mechanical support to the cells.29 Clevers and his group pioneered the development of organoids by using single mouse intestinal ASCs under specific culture conditions that could reproduce the in vivo stem cell niche, thereby inducing the proliferation and differentiation of the intestinal crypt epithelium.102
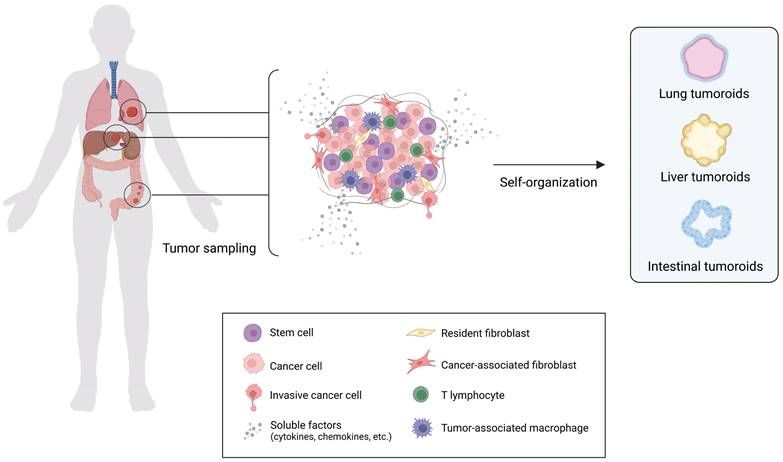
Organoid cultures have been established for several healthy and cancer tissues, such as colon, breast, liver, lung, pancreas, prostate, ovary, among others. 3,103,104 The employment of patient-derived tumor samples has led to the creation of the well-known model commonly referred to as “tumoroid”. 3,103,104
Tumoroids include different cellular subpopulations with specific genetic alterations, thereby maintaining the cancer heterogeneity normally found in vivo. They also retain the genetic signature of the host patient, making them a predictive platform for studying patient response to treatment, guiding the decision-making processes, and improving the effectiveness and efficiency of clinical studies.101 Tumoroids can be developed by engineering healthy organoids using genetic editing technologies such as CRISPR-Cas9, allowing a deep investigation into the onset and progression of tumors as well as testing new therapeutic tools.105 An engineered gastric cancer organoid model with and without AT-rich interactive domain 1A (ARID1A) mutations has been employed to address the context-dependent role for ARID1A in early neoplastic transformation and to identify genotype-dependent therapeutic vulnerabilities. 106 Even more complex models of human colon organoids, recreating the sequence with up to five different oncogenic mutations in APC, KRAS, TP53, SMAD4 and PI3K catalytic subunit-α (PIK3CA), has been utilized for testing drug response. 107,108
Disease modelling in animals is limited by interspecies differences, a limit that can be overcome by human-derived organoids. Additional advantages of organoid cultures include their ability to be expanded in vitro and maintained genetically and phenotypically stable over prolonged periods. Moreover, they can undergo genetic modifications and cryopreservation, facilitating the establishment of an organoid biobank for preclinical studies, encompassing various cancer types directly derived from patients.103,104,109,110 Furthermore, the generation and maintenance of organoids represent significantly more efficient and economically advantageous processes, compared to PDTX models.111
Overall, these characteristics consolidate organoids as an optimal model for personalized cancer medicine.
The ability to model healthy and cancerous tissues simultaneously from the same patient stands out as an important advantage of organoids, providing an effective tool for drug screening. This model aids in the identification of compounds that selectively target cancer cells over healthy ones, allowing the selection of less toxic substances and, consequently, reducing the risk of side effects.103,112 Additionally, the development of organoids from different tumor regions of the same patient enables the reproduction of the tumor heterogeneity paving the way for a personalized therapy.42 In 2015, van de Wetering and colleagues established the first organoid biobank originating from colorectal cancer patients. The biobank included 20 primary tumors together with organoid cultures derived from adjacent normal tissues. By performing a high-throughput automated drug screening, they evaluated several different compounds, including conventional chemotherapeutics and novel targeted inhibitors, on the complete organoid panel. Subsequently, the drug sensitivity data were correlated with the genomic features of cancers to identify molecular signatures and clinically relevant biomarkers associated with treatment responses.113 In another study, Pauli and collaborators generated tumoroids and corresponding PDTX models from patients affected by different malignancies. Their comparative study revealed histopathological similarities between the latter models and their parental tumors, further validated through whole-exome sequencing.114 The analysis demonstrated that tumoroids preserved the genomic alterations of the tissue of origin during prolonged culture. Furthermore, genome sequencing of numerous tumor samples showed that 85.8% of cases had non-targetable somatic alterations in cancer genes, 9.6% could be affected by off-label drugs, and only 0.4% of identified somatic alterations were susceptible to FDA-approved drugs.114 These findings highlight the potential of reliable cancer models in unveiling new therapeutic options. Screening of 160 drugs, including FDA-approved chemotherapeutics and targeted compounds, showed comparable drug responses between tumoroids and PDTX models.114 Moreover, to comprehensively encompass the diversity of breast cancer, a biobank comprising over 100 organoids derived from both primary and metastatic breast cancers has been established.115 These tumoroids faithfully mimic the typical morphology and histopathology of breast cancer while largely conserving the hormone receptor and Her2 status of the parental tumors. This facilitated in vitro drug screening that corresponded to patient responses.115 Finally, among the various successfully developed biobanks, two organoid platforms have been established, derived from multiple stages and subtypes of ovarian cancer, accurately reflecting intra- and inter-patient heterogeneity. The pharmacological analysis conducted on these organoids included both chemotherapeutics (platinum/taxanes) and targeted agents (PI3K/AKT/mTOR inhibitors or PARP inhibitors), revealing significant differences in drug sensitivity that strongly correlated with clinical responses.116,117
Organoids faithfully replicate the genomic and transcriptomic profiles of the patient, aiding in the identification of potential prognostic biomarkers. In a study, bladder cancer organoids exhibited high concordance with the mutational profiles of the parental tumors, including mutations in genes such as TP53, RB1, FGFR3, or epigenetic regulators, like ARID1A, KMT2C, KMT2D, and KDM6A.118 In another study, early cultures of liver tumoroids (less than 2 months) maintained approximately 92% of each patient's genetic variants, with over 80% of the genetic variants retained even in more advanced organoid cultures (beyond 4 months).119 Additionally, a comparison of the transcriptomes of all primary liver cancer organoids with those derived from healthy livers resulted in the identification of novel prognostic markers. Specifically, this analysis revealed that the overexpression of C19ORF48, DTYMK, or UBE2S in hepatocellular carcinoma and the overexpression of C1QBP in cholangiocytes were associated with an unfavorable prognosis.119 Another study highlighted a high concordance, even after long-term culture, in terms of genomic and transcriptomic profiles between gastric tumoroids, encompassing different subtypes, and the corresponding in vivo tumor tissues.120 In addition to the genomic signature, tumoroids faithfully conserve the specific epigenomic features of the modeled tumor type.111 The analysis of the DNA methylation profile in colon organoids cultured in vitro for a prolonged period (12-14 months) revealed the occurrence of spontaneous promoter hyper-methylation, reminiscent of an aging-like process.121 This epigenetic modification resulted in the silencing of pivotal genes in the Wnt signaling pathway, subsequently leading to a progenitor-like cellular state susceptible to neoplastic transformation through the BrafV600E mutation. In contrast, the absence of such promoter hyper-methylation in short-term cultured organoids resulted in significantly greater resistance to BrafV600E-induced transformation.121 Indeed, the transformation of long-term cultured organoids took only two weeks, while five months were required for the younger organoids. It is noteworthy that the CRISPR/Cas9-mediated editing of important genes in the Wnt signaling pathway, targeted by DNA hypermethylation, successfully reproduces the aging-like spontaneous epigenetic silencing.121
Organoids also stand out as a valuable in vitro platform for exploring the expression profile and the functional role of miRNAs. In a study that examined the role of miRNAs associated with the early stages of tumorigenesis in murine intestinal tumor organoids, microarray analyses revealed a pronounced downregulation of specific miRNAs, such as miR-194 and miR-215, in intestinal tumor organoids compared to those derived from normal intestinal epithelium. Specifically, the enforced expression of miR-194 resulted in the inhibition of a key positive regulator of the cell cycle, E2F3, thereby suppressing the growth of intestinal tumor organoids.122 Furthermore, the forced expression of miR-215 was able to suppress the cancer stem cell signature by decreasing the levels of intestinal stem cell markers, including LGR5.122
Another possible application of organoids is the study of host/pathogen interactions. Clevers and his group has implemented a 3D model to assess the oncogenic potential of Helicobacter pylori in human and murine gastric organoids. This platform can be adapted to other tumors and different bacteria, viruses or parasites, thus allowing in vitro investigation of infection-induced changes in primary cells. 123 Rao and co-workers reported a 3D organoid platform to assess the effects of HBV infection on liver tumorigenesis. Liver organoids retain in vivo features and are able to support the complete HBV replication cycle, thus paving the way for design novel HBV-target therapies. 124 Another study by Toyohara and colleagues established squamocolumnar junction organoids to study the molecular mechanisms involved in HPV18-related cervical carcinogenesis. This model allows to identify novel genes involved in HPV18 early promoter activities, which might serve as therapeutic targets in HPV18-infected cervical lesions. 125
Although organoids have gained considerable importance in both basic and translational cancer research, there are still technical and scientific challenges that need to be addressed to fully exploit their potential. Firstly, pronounced variability exists in the success rates of organoid culture, both across different cancer subtypes and within distinct samples of the same tumor type. Organoids modeling, particularly for specific tumor types, can pose intricate challenges. Secondly, different studies on cancer organoids introduce substantial technical variability due to the use of non-standardized and inadequately defined culture protocols, including variation in the source of tumor tissue, culture medium formulations, and the use of animal-derived 3D matrices. This variability, in turn, translates into an inaccurate representation of the biological heterogeneity of cancer, potentially affecting drug development and the identification of biomarkers. Thirdly, the initial growth of non-malignant epithelial cells, identified and confirmed solely through genomic sequencing, must be considered. Lastly, tumoroids fail to capture the TME, as they often exclusively include cancer cells and lack other essential cell types such as fibroblasts, immune cells, and endothelial cells. These cells regulate biological processes like cell proliferation, ECM production, vascularity, angiogenesis, and anti-tumor immunity, driving drug response and tumor aggressiveness. However, this limitation could be overcome through the establishment of a co-culture system.3,40,101,111,126 Colorectal cancer organoids, when cultured in isolation, display a deficiency in gene expression associated with cell-to-cell communication with the TME, a crucial feature present in the cancer tissues of origin. However, upon co-culturing colorectal cancer organoids with CAFs, a patient-dependent re-expression is observed in various genes originally present in the tumor tissue and recognized for their oncogenic functions. This serves as compelling evidence that the microenvironment actively contributes to the regulation of processes involved in tumor progression, such as differential gene expression.127
Another important point to consider is the difficulties to completely reproduce within the tumoroid the complex dynamic of the immune environment. This is relevant in view of the importance that immunotherapy has nowadays. 128 However, attempts in this sense are being made by co-culturing organoids with peripheral blood mononuclear cells (PBMCs) or immune cells from lymph nodes and CAFs to create a TME in which tumor cells are embedded in cytokines and all types of immune cells. 129
Moving forward, there are several opportunities to develop more sophisticated models that can better mimic in vivo conditions. Tumor organoids have been integrated with advanced technologies such as 3D-bioprinting and/or microfluidic chip systems. The combination of microfluidic devices with organoids results in “organoids-on-a-chip”, offering a unique approach to investigate tumor-stroma interactions and their systemic effects. On other hand, combining 3D-bioprinting with organoids ensures the proper spatial arrangement of cells within intricate 3D constructs and preserves the hierarchical architecture of the TME, and enhances model reproducibility.15
Exploring the Potential of Microfluidics in Clinical Research and Medicine
Undoubtedly the 3D in vitro culture systems helped to bridge the gap between traditional 2D cell cultures and animal models by exploiting the strengths of the two approaches. While 3D models strike a balance between ease of manipulation and closer representation of in vivo physiology, they still fall short of completely replicating the in vivo scenario. This limitation is attributed to the absence of native physiological stimuli that cells experience in their living microenvironment.
Within 3D tissues, a network of arteries, veins, and capillaries ensures continuous perfusion of oxygen and nutrients, as well as of drugs, throughout entire organs. The spatial organization plays a crucial role, exposing cell populations to varying gradients of substances that significantly influence the phenotypic and metabolic behavior of the tissues.46,130 Fluid flow is thus considered the part and the parcel of the tissue architecture. The fluidic system, influenced by vessel type, tissue structure, and organism size, generates physiological shear stress impacting the morphology, viability, proliferation, differentiation, and gene expression of a cell and its interactions with other cell populations. These effects are observed not only under normal conditions but also in pathological states.131-133 Immune cells exploit the systemic circulation to identify the regions of the organism affected by inflammation or damage, with their extravasation typically occurring in the vessel walls experiencing higher shear stress.134 Additionally, shear stress plays a pivotal role in regulating cancer invasion and metastasis spread. On one hand, cancer cells utilize systemic circulation to disseminate and colonize distant organs, especially during intra- and extravasation processes. On the other hand, the shear forces within the vessels may hamper the survival of invading cancer cells, hindering their successful metastasis.131,135
Organization of Tumor Organoids. Schematic representation of in vitro heterotypic organoids (refer as tumoroids) formation. Tumoroids are simplified versions of the patient-derived tumor mass containing stem cells that drive the self-organization in the 3D architectures capable of recapitulating several aspects of the complexity and functionality of the corresponding in vivo tissue (created with BioRender).
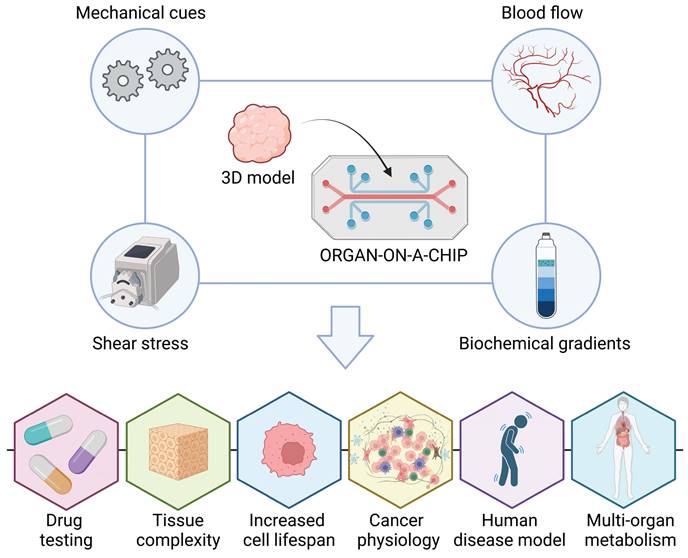
This evidence highlights the importance of the fluid dynamics in the biological systems. Therefore, to address the above-mentioned limitations of 3D static conditions, while adhering to the 3Rs' principles, microfluidic devices have recently gained widespread application in clinical research.136 This promising solution involves integrating 3D models with microfluidics, giving rise to the concept of an "organ-on-a-chip” (Figure 4). This system provides a practical platform for simulating in vivo body fluid perfusion, enabling to mimic the blood flow in organs through externally controlled microfluidics, facilitating the recreation of biochemical gradients and mechanical cues.137 They enable the reconstruction of 3D living tissues under micro physiological fluid-dynamic conditions, facilitated by a peristaltic pump that replicates the speed, direction, and shear stress of fluid flow. This technology recapitulates diverse flows and tissue complexity 132,138, enhancing the reproducibility of tissue conditions, extending cell lifespan, accelerating drug testing without involving animal models, and achieving reliable human disease models for basic research.
Organ-on-a-chip technologies enable the comprehensive study of various aspects of cancer physiology. This includes the recreation of the TME, investigation of cancer-stroma and -immune crosstalk 139, analysis of cell migration and metastasis, screening of anticancer drugs, prediction of therapy responses, and exploration of the transport of anticancer nanomedicines in tumor tissues 140. As an example, creating 3D SKOV-3 cell-laden alginate hydrogels as models for ovarian tumors under fluid dynamic conditions provides a more precise representation of the 3D TME and improves the prediction of in vivo drug efficacy compared to static in vitro models and xenograft mouse models.130 After a week of treating 3D hydrogels with 10 μM cisplatin, cell viability exceeded 80% under static conditions but declined by up to 50% in dynamic culture, which reflected the different cisplatin diffusion rate in the two conditions. Notably, in a xenograft model, the drug efficacy test demonstrated around 44% tumor regression after 5 weeks, aligning with predictions from shorter-term fluid-dynamic in vitro tests.130
The immune-organ-on-a-chip replicates the infiltration of circulating immune cells into a 3D tumor model, offering dual access to both tumor and circulating compartments. This enables the monitoring and quantification of changes in the TME, including soluble molecules, cell death, and tumor cell invasion. Within the microfluidic device, circulating NK cells undergo a spontaneous extravasation process, retaining their ability to interact with matrix-embedded neuroblastoma cells, thereby exhibiting a cytotoxic effect that leads to tumor cell apoptosis.141
Microfluidic devices are also employed in studying the metastatic dissemination of tumor cells, particularly for isolating circulating tumor cells (CTCs). The detection of CTCs serves as a predictive measure for tumor staging. CTC isolation utilizes label-free filtering methods 142,143 or label-based approaches involving specific antibodies.144 Researchers have replicated a 3D circulation system with different patterns of wall shear stress to investigate its effects on the behavior of circulating metastatic breast cancer cells injected into the device. 131
Organ-on-a-chip devices could also facilitate the intricate dynamics of multi-organ metabolism when interconnected in various organs-on-a-chip, as well as the pharmacokinetics 145, including the mechanism of action and toxic effects of drugs 146. A multi-organ chip was used to interconnect ovarian cancer tissues with hepatic cellular models, emulating systemic cisplatin administration. This setup allowed the simultaneous evaluation of drug efficacy and hepatotoxic effects in a physiological context. Notably, the combination of 3D culture, fluid-dynamic conditions, and multi-organ connection showcased superior predictive toxicity and efficacy results compared to clinical therapy. 147
In addition, microfluidic-coupled devices represent a valuable platform to be exploited in precision medicine, particularly for the preclinical screening of anticancer nano-drugs and nanotheranostic systems. 148
Advancing Cancer Research through 3D-Bioprinting for Personalized Medicine
Despite significant progress in 3D co-culture and microfluidics, fully capturing the complexity of in vivo tumors remains a major challenge. The TME consists of a complex array of elements, including the ECM, stromal cells, immune cells, soluble mediators, and blood vessels. 3D-bioprinting enables the precise replication of this intricate architecture by creating tissues and organs that faithfully reproduce the cellular density, ECM composition, and three-dimensional spatial organization of the TME. 15 In recent years, 3D-bioprinting has emerged as an innovative approach for fabricating complex tissue models by producing biomaterials through designed structures. These biomaterials possess powerful properties such as biocompatibility, controllability, printability, and crosslinking, enabling diverse applications in regenerative medicine, cancer research, drug discovery, toxicology, and basic research. 149 For the successful implementation of 3D-bioprinting technology, the use of bioinks is fundamental, as they have recently emerged as indispensable for achieving fast and dependable 3D-bioprinted cell culture systems. 150 Bioinks consist of biocompatible hydrogels embedded with various forms of living cells, including single cells, cell aggregates in spheroids, and cells organized into organoids. 150,151 These biopolymeric hydrogels should replicate the structural, physicochemical, and biological characteristics of the ECM, enhancing the simultaneous attachment and proliferation of various cell types. 152 Additionally, bioinks may include biomolecules such as growth factors, DNA, miRNA, cytokines, and exosomes. 150 By using the bioprinter, hydrogels, cells, and biomolecules can be precisely layered and arranged to control the spatial placement of functional components, enabling the creation of customized 3D structures. 153
Microfluidic Devices: Advancing 3D Tissue Models Towards Organ-on-a-Chip Technology. Microfluidic devices have recently gained widespread application in clinical research to address the limitations of static 3D conditions while adhering to the principles of the 3Rs. 3D models with microfluidics have led to the development of an “organ-on-a-chip concept”. These systems provide a practical platform for simulating in vivo body fluid perfusion, mimicking organ blood flow through externally controlled microfluidics. This capability facilitates the recreation of biochemical gradients and mechanical cues essential for tissue function. “Organ-on-a-chip” enhances reproducibility of tissue conditions, extends cell lifespan, accelerates drug testing without animal models, and establishes reliable human disease models for basic research (created with BioRender).
The combination of organoids with 3D-bioprinting offers a promising avenue for developing more advanced and accurate cancer models, focusing on clinical applications such as chemotherapeutic drug screening and the development of personalized treatment regimens for cancer patients. 15,61
Bioprinted cancer models integrating patient-derived cancer cells and stroma components to mimic the TME and vascularization offer advanced platforms for precision chemotherapy screening across various cancers. For example, Han et al. developed a bioprinting method to recreate the TME by printing a layer of blood vessels using fibroblasts and endothelial cells within a biocompatible matrix. The glioblastoma tumor spheroids were added, and the formation of blood vessel sprouts around the spheroids was observed, which increased their size. Treatment with the drug temozolomide effectively reduced spheroid growth, demonstrating that this bioprinted model is a valuable tool for studying tumor biology and testing drug efficacy. 154 A high throughput bioprinting platform using tunable 3D hydrogels proved effective for testing anti-metastatic drugs toward cancer cells exhibiting distinct migratory and invasive behaviors in dependence of the hydrogel stiffness. 155
Altogether, these models hold promise for personalized medicine by closely mimicking in vivo conditions and improving treatment strategies. 156
Furthermore, the integration of 3D-bioprinting with innovative technologies, such as organ-on-a-chip systems, is likely to revolutionize cancer research. Organ-on-a-chip devices, which mimic the physiological functions of human organs as mentioned above, can be combined with bioprinted tissues to create highly dynamic and interactive cancer models. 157,158
The continuous evolution of 3D-bioprinting technology offers a powerful means to replicate the intricate features of the TME, bringing us closer to creating more accurate cancer models. This progress aligns with the growing demand for prognostic preclinical models that identify the most suitable treatment regimens for individual patients, ultimately enhancing treatment efficacy and minimizing adverse effects, and thus advancing the field of precision medicine.
Concluding Remarks
This review provides a comprehensive overview of primary models employed in cancer research, including in vitro cell cultures and animal models. Particular emphasis is placed on their distinctive characteristics, strengths, and limitations. Here, we meticulously examined the 3D in vitro cell cultures of spheroids and organoids in all their variant implementations, including microfluidic and bioprinting. We stress how these 3D models may represent valid alternatives to the oversimplified and unrealistic 2D in vitro models as well as to animal models, that cannot accurately replicate the complex human conditions. We have thoroughly discussed the advantages offered by tumor spheroids and organoids in comparison to 2D and animal models, particularly in drug screening, differential gene and protein expression, epigenetic regulation, cellular signaling, and biomarkers identification.
In our discussion, we highlight the strengths of 3D models, while acknowledging their limitations as summarized in Table 1.
Overview of the main advantages and limitations of 3D models discussed in the review. Abbreviations: ECM, extracellular matrix; TME, tumor microenvironment.
| Experimental model | PROS | CONS |
|---|---|---|
| Spheroids | Low expensive and time effective (scalable in high throughput) Easy manipulation and high accessibility of cell materials High reproducibility and long-term growth Simplicity of genetic manipulation Possibility of setting up co-cultures for mimicking tumor heterogenicity | Oversimplified static system Lack of robustness and reliability for translational relevance Lack of ability to recapitulate in vivo organ features |
| Organoids | Faithfully recapitulate the pathophysiology of the disease Highly predictive platform for studying patient responses Accurately replicate tissue architecture, functionality and dynamic complexity of TME Simplicity of genetic manipulation Possibility of setting up co-cultures for mimicking tumor heterogenicity Possibility to model healthy and tumor tissues simultaneously from the same patient Development of biobanks | Challenging manipulation Need of ethical approval for patients-derived samples High expensive Limited growth (depending on stemness/differentiation ratio) Limited reproducibility Oversimplified static system |
| Organ-on-chip | Combination of advantages of microfluidic devices with organoids Possibility of interconnecting different organoids to recreate multi-organ metabolism (multi-organ-on-chip) | High expensive Require expertise Need of ethical approval for patients-derived samples Difficult to adapt to high throughput Readout typically limited to end-point analysis |
| Microfluidics | Replicate physiological TME conditions, vasculature-like perfusion, precise control over chemical gradient flows, and mechanical forces Possibility of collecting the fluids for characterization of secreted soluble factors and circulating cells Implementation of a dynamic system | Low amount of samples available for downstream characterizations Require expertise High expensive Difficult to adapt to high throughput |
| 3D-Bioprinting | Faithfully recapitulate complex architecture, precise control over chemical gradient flows, and mechanical forces (stiffness) of native ECM Proper spatial arrangement of cells within intricate 3D constructs and preserves the hierarchical architecture of the TME High reproducibility | Require disruption of hydrogel/matrix to collect cells Low amount of samples available for downstream characterizations Require expertise High expensive Difficult to adapt to high throughput |
Overview of the Current Applications of 3D Models in Cancer Research. Schematic representation of the suitability of each 3D culture system (spheroids, organoids, organ-on-chip, microfluidics and 3D-Bioprinting) to study the onset and progression of carcinogenesis, efficacy of anticancer therapies, cell-cell crosstalk in the TME, host/pathogen or host/microbiota interplays, expression profiling, and metastatic process (created with Biorender).
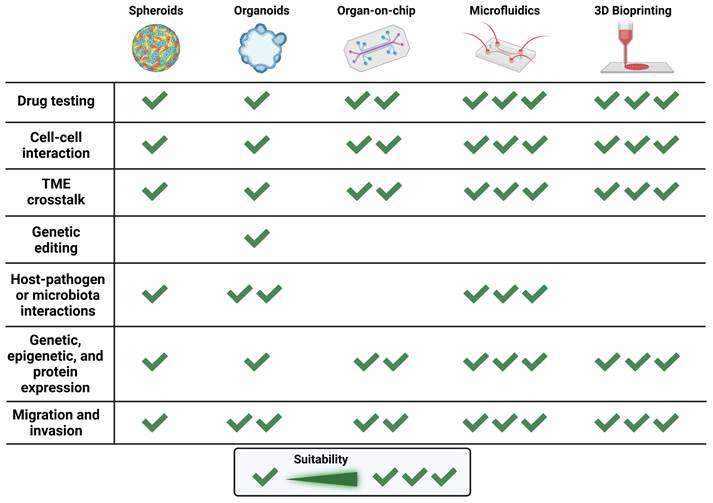
The implementation with microfluidics holds promising potential in 3D models exploited in clinical research. Further investigations are essential to uncover new possibilities in microfluidic applications, potentially leading to more personalized and precise medicine. This evolving field has the capacity to drive breakthroughs in diagnostics, drug delivery, and tissue engineering, offering innovative solutions to complex challenges in diverse medical fields. A brief overview of the possible applications and advantages of all the 3D models mentioned above are illustrated in Figure 5.
Taken together, 3D models emerge as a pivotal tool bridging the gap between 2D in vitro and in vivo models. The increasing complexity of 3D cell cultures, resulted from the integration of spheroids and organoids with microfluidic systems and/or 3D-bioprinting, brings researchers closer to mimicking in vivo conditions. The employment of 3D models represents a significant advancement in in vitro research, contributing to the development of effective antitumor strategies. Their application promises a reduction in animal experimentation, offering advantages in terms of costs, time efficiency, and ethical considerations. Consequently, this advancement enhances the robustness and reliability of research data, facilitating the seamless translation of findings from the experimental phase to clinical applications. We are aware that in vitro models, although sophisticated and useful for understanding biological mechanisms, also present important limitations as they cannot reveal organ toxicity or provide information on pharmacokinetics, which can be derived from animal studies.21 Here we presented the bioengineered 3D in vitro models that could serve as valid alternative platforms to limit the use of animal models for preclinical testing.
Acknowledgements
A.E. is recipient of a PhD fellowship granted by the Italian Ministry of Education, University and Research (MIUR, Rome, Italy). A.F. is recipient of a post-doctoral fellowship from Fondazione Umberto Veronesi (FUV2024). L.V. is recipient of a post-doctoral fellowship granted by the Università del Piemonte Orientale (Novara, Italy). B.G. is recipient of a PhD fellowship granted by Comoli, Ferrari & SpA (Novara, Italy). Authors are grateful for the support of the Consorzio Interuniversitario per le Biotecnologie (CIB, Trieste, Italy).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023Jan;73(1):17-48 doi: 10.3322/caac.21763. PMID: 36633525
2. Jubelin C, Muñoz-Garcia J, Griscom L, Cochonneau D, Ollivier E, Heymann MF, Vallette FM, Oliver L, Heymann D. Three-dimensional in vitro culture models in oncology research. Cell Biosci. 2022Sep11;12(1):155 doi: 10.1186/s13578-022-00887-3. PMID: 36089610; PMCID: PMC9465969
3. Zanoni M, Cortesi M, Zamagni A, Arienti C, Pignatta S, Tesei A. Modeling neoplastic disease with spheroids and organoids. J Hematol Oncol. 2020Jul16;13(1):97 doi: 10.1186/s13045-020-00931-0. PMID: 32677979; PMCID: PMC7364537
4. Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M, White C, Lowe C, Sherba JJ, Hartmanshenn C, O'Neill KM, Balter ML, Fritz ZR, Androulakis IP, Schloss RS, Yarmush ML. The growing role of precision and personalized medicine for cancer treatment. Technology (Singap World Sci). 2018;6:79-100 doi: 10.1142/S2339547818300020. Epub 2019 Jan 11. PMID: 30713991; PMCID: PMC6352312
5. Prasad V, Mailankody S. Research and Development Spending to Bring a Single Cancer Drug to Market and Revenues After Approval. JAMA Intern Med. 2017 Nov 1;177(11):1569-1575. doi: 10.1001/jamainternmed.2017.3601. Erratum in: JAMA Intern Med. 2017 Nov 1;177(11):1703. Erratum in: JAMA Intern Med. 2018Oct1;178(10):1433 PMID: 28892524; PMCID: PMC5710275
6. Amirghasemi F, Adjei-Sowah E, Pockaj BA, Nikkhah M. Microengineered 3D Tumor Models for Anti-Cancer Drug Discovery in Female-Related Cancers. Ann Biomed Eng. 2021Aug;49(8):1943-1972 doi: 10.1007/s10439-020-02704-9. Epub 2021 Jan 5. PMID: 33403451
7. Franchi-Mendes T, Eduardo R, Domenici G, Brito C. 3D Cancer Models: Depicting Cellular Crosstalk within the Tumour Microenvironment. Cancers (Basel). 2021Sep14;13(18):4610 doi: 10.3390/cancers13184610. PMID: 34572836; PMCID: PMC8468887
8. Barbosa MAG, Xavier CPR, Pereira RF, Petrikaitė V, Vasconcelos MH. 3D Cell Culture Models as Recapitulators of the Tumor Microenvironment for the Screening of Anti-Cancer Drugs. Cancers (Basel). 2021Dec31;14(1):190 doi: 10.3390/cancers14010190. PMID: 35008353; PMCID: PMC8749977
9. Moreno L, Pearson AD. How can attrition rates be reduced in cancer drug discovery? Expert Opin Drug Discov. 2013Apr;8(4):363-8 doi: 10.1517/17460441.2013.768984. Epub 2013 Feb 4. PMID: 23373702
10. Nunes AS, Barros AS, Costa EC, Moreira AF, Correia IJ. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng. 2019Jan;116(1):206-226 doi: 10.1002/bit.26845. Epub 2018 Oct 27. PMID: 30367820
11. Roy V, Magne B, Vaillancourt-Audet M, Blais M, Chabaud S, Grammond E, Piquet L, Fradette J, Laverdière I, Moulin VJ, Landreville S, Germain L, Auger FA, Gros-Louis F, Bolduc S. Human Organ-Specific 3D Cancer Models Produced by the Stromal Self-Assembly Method of Tissue Engineering for the Study of Solid Tumors. Biomed Res Int. 2020Apr13;2020:6051210 doi: 10.1155/2020/6051210. PMID: 32352002; PMCID: PMC7178531
12. Pozzi S, Scomparin A, Israeli Dangoor S, Rodriguez Ajamil D, Ofek P, Neufeld L, Krivitsky A, Vaskovich-Koubi D, Kleiner R, Dey P, Koshrovski-Michael S, Reisman N, Satchi-Fainaro R. Meet me halfway: Are in vitro 3D cancer models on the way to replace in vivo models for nanomedicine development? Adv Drug Deliv Rev. 2021Aug;175:113760 doi: 10.1016/j.addr.2021.04.001. Epub 2021 Apr 7. PMID: 33838208
13. Hoarau-Véchot J, Rafii A, Touboul C, Pasquier J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int J Mol Sci. 2018Jan18;19(1):181 doi: 10.3390/ijms19010181. PMID: 29346265; PMCID: PMC5796130
14. Bielecka ZF, Maliszewska-Olejniczak K, Safir IJ, Szczylik C, Czarnecka AM. Three-dimensional cell culture model utilization in cancer stem cell research. Biol Rev Camb Philos Soc. 2017Aug;92(3):1505-1520 doi: 10.1111/brv.12293. Epub 2016 Aug 22. PMID: 27545872
15. Rodrigues J, Heinrich MA, Teixeira LM, Prakash J. 3D In Vitro Model (R)evolution: Unveiling Tumor-Stroma Interactions. Trends Cancer. 2021Mar;7(3):249-264 doi: 10.1016/j.trecan.2020.10.009. Epub 2020 Nov 18. PMID: 33218948
16. Hynds RE, Vladimirou E, Janes SM. The secret lives of cancer cell lines. Dis Model Mech. 2018Nov16;11(11):dmm037366 doi: 10.1242/dmm.037366
17. Li Z, Zheng W, Wang H, Cheng Y, Fang Y, Wu F, Sun G, Sun G, Lv C, Hui B. Application of Animal Models in Cancer Research: Recent Progress and Future Prospects. Cancer Manag Res. 2021Mar15;13:2455-2475 doi: 10.2147/CMAR.S302565. PMID: 33758544; PMCID: PMC7979343
18. Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA; Committee of the National Cancer Research Institute. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010May25;102(11):1555-77 doi: 10.1038/sj.bjc.6605642. PMID: 20502460; PMCID: PMC2883160
19. Londoño-Berrio M, Castro C, Cañas A, Ortiz I, Osorio M. Advances in Tumor Organoids for the Evaluation of Drugs: A Bibliographic Review. Pharmaceutics. 2022Dec3;14(12):2709 doi: 10.3390/pharmaceutics14122709. PMID: 36559203; PMCID: PMC9784359
20. Chuprin J, Buettner H, Seedhom MO, Greiner DL, Keck JG, Ishikawa F, Shultz LD, Brehm MA. Humanized mouse models for immuno-oncology research. Nat Rev Clin Oncol. 2023Mar;20(3):192-206 doi: 10.1038/s41571-022-00721-2. Epub 2023 Jan 12. PMID: 36635480; PMCID: PMC10593256
21. Karnik I, Her Z, Neo SH, Liu WN, Chen Q. Emerging Preclinical Applications of Humanized Mouse Models in the Discovery and Validation of Novel Immunotherapeutics and Their Mechanisms of Action for Improved Cancer Treatment. Pharmaceutics. 2023May26;15(6):1600 doi: 10.3390/pharmaceutics15061600. PMID: 37376049; PMCID: PMC10305679
22. Tian H, Lyu Y, Yang YG, Hu Z. Humanized Rodent Models for Cancer Research. Front Oncol. 2020Sep11;10:1696 doi: 10.3389/fonc.2020.01696. PMID: 33042811; PMCID: PMC7518015
23. Brancato V, Oliveira JM, Correlo VM, Reis RL, Kundu SC. Could 3D models of cancer enhance drug screening? Biomaterials. 2020Feb;232:119744 doi: 10.1016/j.biomaterials.2019.119744. Epub 2019 Dec 26. PMID: 31918229
24. Radhakrishnan J, Varadaraj S, Dash SK, Sharma A, Verma RS. Organotypic cancer tissue models for drug screening: 3D constructs, bioprinting and microfluidic chips. Drug Discov Today. 2020May;25(5):879-890 doi: 10.1016/j.drudis.2020.03.002. Epub 2020 Mar 9. PMID: 32165322
25. Seyhan AA. Lost in translation: the valley of death across preclinical and clinical divide - identification of problems and overcoming obstacles. Transl Med Commun. 2019;4:43 https://doi.org/10.1186/s41231-019-0050-7
26. Steimberg N. et al. In Vitro Modeling of Tissue-Specific 3D Microenvironments and Possibile Application to Pediatric Cancer Research. Journal of Pediatric Oncology. 2014;2:40-76 10.14205/2309-3021.2014.02.01.5
27. Caballero D. et al. Forecast cancer: the importance of biomimetic 3D in vitro models in cancer drug testing/discovery and therapy. In Vitro Models (Springer). 2022;1:119-123 10.1007/s44164-022-00014-z
28. Suarez-Martinez E, Suazo-Sanchez I, Celis-Romero M, Carnero A. 3D and organoid culture in research: physiology, hereditary genetic diseases and cancer. Cell Biosci. 2022Apr1;12(1):39 doi: 10.1186/s13578-022-00775-w. PMID: 35365227; PMCID: PMC8973959
29. Goldrick C, Guri I, Herrera-Oropeza G, O'Brien-Gore C, Roy E, Wojtynska M, Spagnoli FM. 3D multicellular systems in disease modelling: From organoids to organ-on-chip. Front Cell Dev Biol. 2023Feb2;11:1083175 doi: 10.3389/fcell.2023.1083175. PMID: 36819106; PMCID: PMC9933985
30. Sutherland RM, McCredie JA, Inch WR. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J Natl Cancer Inst. 1971Jan;46(1):113-20 PMID: 5101993
31. Costa EC, Moreira AF, de Melo-Diogo D, Gaspar VM, Carvalho MP, Correia IJ. 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnol Adv. 2016Dec;34(8):1427-1441 doi: 10.1016/j.biotechadv.2016.11.002. Epub 2016 Nov 11. PMID: 27845258
32. Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015Jan;17(1):1-15 doi: 10.1016/j.neo.2014.12.004. PMID: 25622895; PMCID: PMC4309685
33. Reynolds DS, Tevis KM, Blessing WA, Colson YL, Zaman MH, Grinstaff MW. Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment. Sci Rep. 2017Sep4;7(1):10382 doi: 10.1038/s41598-017-10863-4. PMID: 28871147; PMCID: PMC5583341
34. Kutle I, Polten R, Hachenberg J, Klapdor R, Morgan M, Schambach A. Tumor Organoid and Spheroid Models for Cervical Cancer. Cancers (Basel). 2023Apr27;15(9):2518 doi: 10.3390/cancers15092518. PMID: 37173984; PMCID: PMC10177622
35. Kondo J, Endo H, Okuyama H, Ishikawa O, Iishi H, Tsujii M, Ohue M, Inoue M. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci U S A. 2011Apr12;108(15):6235-40 doi: 10.1073/pnas.1015938108. Epub 2011 Mar 28. PMID: 21444794; PMCID: PMC3076886
36. Gendre DAJ, Ameti E, Karenovics W, Perriraz-Mayer N, Triponez F, Serre-Beinier V. Optimization of tumor spheroid model in mesothelioma and lung cancers and anti-cancer drug testing in H2052/484 spheroids. Oncotarget. 2021Nov23;12(24):2375-2387 doi: 10.18632/oncotarget.28134. PMID: 34853659; PMCID: PMC8629400
37. Giustarini G, Teng G, Pavesi A, Adriani G. Characterization of 3D heterocellular spheroids of pancreatic ductal adenocarcinoma for the study of cell interactions in the tumor immune microenvironment. Front Oncol. 2023Jul14;13:1156769 doi: 10.3389/fonc.2023.1156769. PMID: 37519820; PMCID: PMC10375712
38. Wartenberg M, Gronczynska S, Bekhite MM, Saric T, Niedermeier W, Hescheler J, Sauer H. Regulation of the multidrug resistance transporter P-glycoprotein in multicellular prostate tumor spheroids by hyperthermia and reactive oxygen species. Int J Cancer. 2005;113(2):229-40 doi: 10.1002/ijc.20596. PMID: 15389514
39. Martinelli S, Cantini G, Propato AP, Bani D, Guasti D, Nardini P, Calosi L, Mello T, Bechmann N, Danza G, Villanelli F, Canu L, Maggi M, Mannelli M, Rapizzi E, Luconi M. The 3D in vitro Adrenoid cell model recapitulates the complexity of the adrenal gland. Sci Rep. 2024Apr5;14(1):8044 doi: 10.1038/s41598-024-58664-w. PMID: 38580769; PMCID: PMC10997590
40. Manduca N, Maccafeo E, De Maria R, Sistigu A, Musella M. 3D cancer models: One step closer to in vitro human studies. Front Immunol. 2023Apr11;14:1175503 doi: 10.3389/fimmu.2023.1175503. PMID: 37114038; PMCID: PMC10126361
41. Lee KH, Kim TH. Recent Advances in Multicellular Tumor Spheroid Generation for Drug Screening. Biosensors (Basel). 2021Nov11;11(11):445 doi: 10.3390/bios11110445. PMID: 34821661; PMCID: PMC8615712
42. Gunti S, Hoke ATK, Vu KP, London NR Jr. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers (Basel). 2021Feb19;13(4):874 doi: 10.3390/cancers13040874. PMID: 33669619; PMCID: PMC7922036
43. Białkowska K, Komorowski P, Bryszewska M, Miłowska K. Spheroids as a Type of Three-Dimensional Cell Cultures-Examples of Methods of Preparation and the Most Important Application. Int J Mol Sci. 2020Aug28;21(17):6225 doi: 10.3390/ijms21176225. PMID: 32872135; PMCID: PMC7503223
44. Cui X, Hartanto Y, Zhang H. Advances in multicellular spheroids formation. J R Soc Interface. 2017Feb;14(127):20160877 doi: 10.1098/rsif.2016.0877. PMID: 28202590; PMCID: PMC5332573
45. Han SJ, Kwon S, Kim KS. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021Mar4;21(1):152 doi: 10.1186/s12935-021-01853-8. PMID: 33663530; PMCID: PMC7934264
46. Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014May;12(4):207-18 doi: 10.1089/adt.2014.573. PMID: 24831787; PMCID: PMC4026212
47. Sharma K, Dey S, Karmakar R, Rengan AK. A comprehensive review of 3D cancer models for drug screening and translational research. Cancer Innov. 2023Dec22;3(1):e102 doi: 10.1002/cai2.102. PMID: 38948533; PMCID: PMC11212324
48. Li W, Hu X, Yang S, Wang S, Zhang C, Wang H, Cheng YY, Wang Y, Liu T, Song K. A novel tissue-engineered 3D tumor model for anti-cancer drug discovery. Biofabrication. 2018Oct30;11(1):015004 doi: 10.1088/1758-5090/aae270. PMID: 30229749
49. Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012Mar20;21(3):418-29 doi: 10.1016/j.ccr.2012.01.007. PMID: 22439937; PMCID: PMC3371414
50. Navab R, Strumpf D, To C, Pasko E, Kim KS, Park CJ, Hai J, Liu J, Jonkman J, Barczyk M, Bandarchi B, Wang YH, Venkat K, Ibrahimov E, Pham NA, Ng C, Radulovich N, Zhu CQ, Pintilie M, Wang D, Lu A, Jurisica I, Walker GC, Gullberg D, Tsao MS. Integrin α11β1 regulates cancer stromal stiffness and promotes tumorigenicity and metastasis in non-small cell lung cancer. Oncogene. 2016Apr14;35(15):1899-908 doi: 10.1038/onc.2015.254. Epub 2015 Jul 6. PMID: 26148229; PMCID: PMC4833874
51. Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011Apr;53(4):1192-205 doi: 10.1002/hep.24108. PMID: 21442631; PMCID: PMC3076070
52. Unnikrishnan K, Thomas LV, Ram Kumar RM. Advancement of Scaffold-Based 3D Cellular Models in Cancer Tissue Engineering: An Update. Front Oncol. 2021Oct25;11:733652 doi: 10.3389/fonc.2021.733652. PMID: 34760696; PMCID: PMC8573168
53. Henke E, Nandigama R, Ergün S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Front Mol Biosci. 2020Jan31;6:160 doi: 10.3389/fmolb.2019.00160. PMID: 32118030; PMCID: PMC7025524
54. Urzì O, Gasparro R, Costanzo E, De Luca A, Giavaresi G, Fontana S, Alessandro R. Three-Dimensional Cell Cultures: The Bridge between In Vitro and In Vivo Models. Int J Mol Sci. 2023Jul27;24(15):12046 doi: 10.3390/ijms241512046. PMID: 37569426; PMCID: PMC10419178
55. Kenny HA, Dogan S, Zillhardt M, K Mitra A, Yamada SD, Krausz T, Lengyel E. Organotypic models of metastasis: A three-dimensional culture mimicking the human peritoneum and omentum for the study of the early steps of ovarian cancer metastasis. Cancer Treat Res. 2009;149:335-51 doi: 10.1007/978-0-387-98094-2_16. PMID: 19763444; PMCID: PMC3651885
56. Canda AE, Sever T, Calibasi Kocal G, Basbinar Y, Ellidokuz H. In vitro 3D microfluidic peritoneal metastatic colorectal cancer model for testing different oxaliplatin-based HIPEC regimens. Pleura Peritoneum. 2024Feb28;9(1):23-29 doi: 10.1515/pp-2023-0033. PMID: 38558874; PMCID: PMC10980980
57. Ren K, Xie X, Min T, Sun T, Wang H, Zhang Y, Dang C, Zhang H. Development of the Peritoneal Metastasis: A Review of Back-Grounds, Mechanisms, Treatments and Prospects. J Clin Med. 2022Dec23;12(1):103 doi: 10.3390/jcm12010103. PMID: 36614904; PMCID: PMC9821147
58. Costa EC, Gaspar VM, Coutinho P, Correia IJ. Optimization of liquid overlay technique to formulate heterogenic 3D co-cultures models. Biotechnol Bioeng. 2014Aug;111(8):1672-85 doi: 10.1002/bit.25210. Epub 2014 Feb 25. PMID: 24615162
59. Martinelli S, Riverso M, Mello T, Amore F, Parri M, Simeone I, Mannelli M, Maggi M, Rapizzi E. SDHB and SDHD silenced pheochromocytoma spheroids respond differently to tumour microenvironment and their aggressiveness is inhibited by impairing stroma metabolism. Mol Cell Endocrinol. 2022May1;547:111594 doi: 10.1016/j.mce.2022.111594. Epub 2022 Feb 9. PMID: 35149119
60. Martinelli S, Amore F, Mello T, Mannelli M, Maggi M, Rapizzi E. Metformin Treatment Induces Different Response in Pheochromocytoma/Paraganglioma Tumour Cells and in Primary Fibroblasts. Cancers (Basel). 2022Jul17;14(14):3471 doi: 10.3390/cancers14143471. PMID: 35884532; PMCID: PMC9320533
61. Cadamuro F, Nicotra F, Russo L. 3D printed tissue models: From hydrogels to biomedical applications. J Control Release. 2023Feb;354:726-745 doi: 10.1016/j.jconrel.2023.01.048. Epub 2023 Jan 26. PMID: 36682728
62. Magliaro C, Mattei G, Iacoangeli F, Corti A, Piemonte V, Ahluwalia A. Oxygen Consumption Characteristics in 3D Constructs Depend on Cell Density. Front Bioeng Biotechnol. 2019Oct10;7:251 doi: 10.3389/fbioe.2019.00251. PMID: 31649925; PMCID: PMC6796794
63. Boucherit N, Gorvel L, Olive D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front Immunol. 2020Dec10;11:603640 doi: 10.3389/fimmu.2020.603640. PMID: 33362787; PMCID: PMC7758240
64. Curcio E, Salerno S, Barbieri G, De Bartolo L, Drioli E, Bader A. Mass transfer and metabolic reactions in hepatocyte spheroids cultured in rotating wall gas-permeable membrane system. Biomaterials. 2007Dec;28(36):5487-97 doi: 10.1016/j.biomaterials.2007.08.033. Epub 2007 Sep 18. PMID: 17881050
65. Carlsson J, Acker H. Relations between pH, oxygen partial pressure and growth in cultured cell spheroids. Int J Cancer. 1988Nov15;42(5):715-20 doi: 10.1002/ijc.2910420515. PMID: 3182108
66. Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol. 2010Jul1;148(1):3-15 doi: 10.1016/j.jbiotec.2010.01.012. Epub 2010 Jan 25. PMID: 20097238
67. Secomandi E, Esposito A, Camurani G, Vidoni C, Salwa A, Lualdi C, Vallino L, Ferraresi A, Isidoro C. Differential Competitive Growth of Transgenic Subclones of Neuroblastoma Cells Expressing Different Levels of Cathepsin D Co-Cultured in 2D and 3D in Response to EGF: Implications in Tumor Heterogeneity and Metastasis. Cancers (Basel). 2024Mar29;16(7):1343 doi: 10.3390/cancers16071343. PMID: 38611021; PMCID: PMC11010890
68. Vallino L, Garavaglia B, Visciglia A, Amoruso A, Pane M, Ferraresi A, Isidoro C. Cell-free Lactiplantibacillus plantarum OC01 supernatant suppresses IL-6-induced proliferation and invasion of human colorectal cancer cells: Effect on β-Catenin degradation and induction of autophagy. J Tradit Complement Med. 2023Feb21;13(2):193-206 doi: 10.1016/j.jtcme.2023.02.001. PMID: 36970462; PMCID: PMC10037073
69. Garavaglia B, Vallino L, Ferraresi A, Esposito A, Salwa A, Vidoni C, Gentilli S, Isidoro C. Butyrate Inhibits Colorectal Cancer Cell Proliferation through Autophagy Degradation of β-Catenin Regardless of APC and β-Catenin Mutational Status. Biomedicines. 2022May13;10(5):1131 doi: 10.3390/biomedicines10051131. PMID: 35625868; PMCID: PMC9138675
70. Tian X, Wang W, Zhang Q, Zhao L, Wei J, Xing H, Song Y, Wang S, Ma D, Meng L, Chen G. Hypoxia-inducible factor-1α enhances the malignant phenotype of multicellular spheroid HeLa cells in vitro. Oncol Lett. 2010Sep;1(5):893-897 doi: 10.3892/ol_00000159. Epub 2010 Sep 1. PMID: 22966402; PMCID: PMC3436219
71. Doublier S, Belisario DC, Polimeni M, Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A, Sapino A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: a potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer. 2012Jan4;12:4 doi: 10.1186/1471-2407-12-4. PMID: 22217342; PMCID: PMC3262753
72. Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011Dec;2(12):1117-33 doi: 10.1177/1947601911423654. PMID: 22866203; PMCID: PMC3411127
73. Qin Y, Roszik J, Chattopadhyay C, Hashimoto Y, Liu C, Cooper ZA, Wargo JA, Hwu P, Ekmekcioglu S, Grimm EA. Hypoxia-Driven Mechanism of Vemurafenib Resistance in Melanoma. Mol Cancer Ther. 2016Oct;15(10):2442-2454 doi: 10.1158/1535-7163.MCT-15-0963. Epub 2016 Jul 25. PMID: 27458138; PMCID: PMC5079683
74. Sonveaux P. ROS and radiotherapy: more we care. Oncotarget. 2017May30;8(22):35482-35483 doi: 10.18632/oncotarget.16613. PMID: 28415657; PMCID: PMC5482588
75. Grimes DR, Partridge M. A mechanistic investigation of the oxygen fixation hypothesis and oxygen enhancement ratio. Biomed Phys Eng Express. 2015Dec4;1(4):045209 doi: 10.1088/2057-1976/1/4/045209. PMID: 26925254; PMCID: PMC4765087
76. Kovacic P, Osuna JA Jr. Mechanisms of anti-cancer agents: emphasis on oxidative stress and electron transfer. Curr Pharm Des. 2000Feb;6(3):277-309 doi: 10.2174/1381612003401046. PMID: 10637380
77. Wartenberg M, Hoffmann E, Schwindt H, Grünheck F, Petros J, Arnold JR, Hescheler J, Sauer H. Reactive oxygen species-linked regulation of the multidrug resistance transporter P-glycoprotein in Nox-1 overexpressing prostate tumor spheroids. FEBS Lett. 2005;579(20):4541-4549 doi: 10.1016/j.febslet.2005.06.078. PMID: 16083877
78. Deavall DG, Martin EA, Horner JM, Roberts R. Drug-induced oxidative stress and toxicity. J Toxicol. 2012;2012:645460 doi: 10.1155/2012/645460. Epub 2012 Aug 5. PMID: 22919381; PMCID: PMC3420138
79. Imamura Y, Mukohara T, Shimono Y, Funakoshi Y, Chayahara N, Toyoda M, Kiyota N, Takao S, Kono S, Nakatsura T, Minami H. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol Rep. 2015Apr;33(4):1837-43 doi: 10.3892/or.2015.3767. Epub 2015 Jan 29. PMID: 25634491
80. Cowan DS, Tannock IF. Factors that influence the penetration of methotrexate through solid tissue. Int J Cancer. 2001Jan1;91(1):120-5 doi: 10.1002/1097-0215(20010101)91:1<120::aid-ijc1021>3.0.co;2-y. PMID: 11149410
81. Swietach P, Hulikova A, Patiar S, Vaughan-Jones RD, Harris AL. Importance of intracellular pH in determining the uptake and efficacy of the weakly basic chemotherapeutic drug, doxorubicin. PLoS One. 2012;7(4):e35949 doi: 10.1371/journal.pone.0035949. Epub 2012 Apr 26. PMID: 22563426; PMCID: PMC3338554
82. Wojtkowiak JW, Verduzco D, Schramm KJ, Gillies RJ. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol Pharm. 2011Dec5;8(6):2032-8 doi: 10.1021/mp200292c. Epub 2011 Oct 26. PMID: 21981633; PMCID: PMC3230683
83. Heldin CH, Rubin K, Pietras K, Ostman A. High interstitial fluid pressure - an obstacle in cancer therapy. Nat Rev Cancer. 2004Oct;4(10):806-13 doi: 10.1038/nrc1456. PMID: 15510161
84. Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006Aug;6(8):583-92 doi: 10.1038/nrc1893. PMID: 16862189
85. Green SK, Francia G, Isidoro C, Kerbel RS. Antiadhesive antibodies targeting E-cadherin sensitize multicellular tumor spheroids to chemotherapy in vitro. Mol Cancer Ther. 2004Feb;3(2):149-59 PMID: 14985455
86. Birgersdotter A, Sandberg R, Ernberg I. Gene expression perturbation in vitro-a growing case for three-dimensional (3D) culture systems. Semin Cancer Biol. 2005Oct;15(5):405-12 doi: 10.1016/j.semcancer.2005.06.009. PMID: 16055341
87. Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007Jun;1(1):84-96 doi: 10.1016/j.molonc.2007.02.004. PMID: 18516279; PMCID: PMC2391005
88. Ghosh S, Spagnoli GC, Martin I, Ploegert S, Demougin P, Heberer M, Reschner A. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: a high-density oligonucleotide array study. J Cell Physiol. 2005Aug;204(2):522-31 doi: 10.1002/jcp.20320. PMID: 15744745
89. Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer KL, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, Krieg A, Stoecklein NH. Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One. 2013;8(3):e59689 doi: 10.1371/journal.pone.0059689. Epub 2013 Mar 26. PMID: 23555746; PMCID: PMC3608563
90. Kim H, Phung Y, Ho M. Changes in global gene expression associated with 3D structure of tumors: an ex vivo matrix-free mesothelioma spheroid model. PLoS One. 2012;7(6):e39556 doi: 10.1371/journal.pone.0039556. Epub 2012 Jun 21. PMID: 22737246; PMCID: PMC3380922
91. Chang TT, Hughes-Fulford M. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng Part A. 2009Mar;15(3):559-67 doi: 10.1089/ten.tea.2007.0434. PMID: 18724832; PMCID: PMC6468949
92. Salinas-Vera YM, Valdés J, Pérez-Navarro Y, Mandujano-Lazaro G, Marchat LA, Ramos-Payán R, Nuñez-Olvera SI, Pérez-Plascencia C, López-Camarillo C. Three-Dimensional 3D Culture Models in Gynecological and Breast Cancer Research. Front Oncol. 2022May26;12:826113 doi: 10.3389/fonc.2022.826113. PMID: 35692756; PMCID: PMC9177953
93. Thippabhotla S, Zhong C, He M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep. 2019Sep10;9(1):13012 doi: 10.1038/s41598-019-49671-3. PMID: 31506601; PMCID: PMC6736862
94. Yoshimura A, Sawada K, Nakamura K, Kinose Y, Nakatsuka E, Kobayashi M, Miyamoto M, Ishida K, Matsumoto Y, Kodama M, Hashimoto K, Mabuchi S, Kimura T. Exosomal miR-99a-5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer. 2018Nov5;18(1):1065 doi: 10.1186/s12885-018-4974-5. PMID: 30396333; PMCID: PMC6217763
95. Esposito A, Ferraresi A, Salwa A, Vidoni C, Dhanasekaran DN, Isidoro C. Resveratrol Contrasts IL-6 Pro-Growth Effects and Promotes Autophagy-Mediated Cancer Cell Dormancy in 3D Ovarian Cancer: Role of miR-1305 and of Its Target ARH-I. Cancers (Basel). 2022Apr25;14(9):2142 doi: 10.3390/cancers14092142. PMID: 35565270; PMCID: PMC9101105
96. Nguyen HT, Li C, Lin Z, Zhuang Y, Flemington EK, Burow ME, Lin YI, Shan B. The microRNA expression associated with morphogenesis of breast cancer cells in three-dimensional organotypic culture. Oncol Rep. 2012Jul;28(1):117-126 doi: 10.3892/or.2012.1764. Epub 2012 Apr 19. PMID: 22576799; PMCID: PMC3991116
97. Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M, Kenner L, Pfeiffer D, Krupitza G, Dolznig H. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci. 2017Jan1;130(1):203-218 doi: 10.1242/jcs.188102. Epub 2016 Sep 23. PMID: 27663511
98. Gangadhara S, Smith C, Barrett-Lee P, Hiscox S. 3D culture of Her2+ breast cancer cells promotes AKT to MAPK switching and a loss of therapeutic response. BMC Cancer. 2016Jun1;16:345 doi: 10.1186/s12885-016-2377-z. PMID: 27251376; PMCID: PMC4888214
99. Yang S, Hu H, Kung H, Zou R, Dai Y, Hu Y, Wang T, Lv T, Yu J, Li F. Organoids: The current status and biomedical applications. MedComm (2020). 2023May17;4(3):e274 doi: 10.1002/mco2.274. PMID: 37215622; PMCID: PMC10192887
100. Clevers H. Modeling Development and Disease with Organoids. Cell. 2016Jun16;165(7):1586-1597 doi: 10.1016/j.cell.2016.05.082. PMID: 27315476
101. Novelli G, Spitalieri P, Murdocca M, Centanini E, Sangiuolo F. Organoid factory: The recent role of the human induced pluripotent stem cells (hiPSCs) in precision medicine. Front Cell Dev Biol. 2023Jan9;10:1059579 doi: 10.3389/fcell.2022.1059579. PMID: 36699015; PMCID: PMC9869172
102. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009May14;459(7244):262-5 doi: 10.1038/nature07935. Epub 2009 Mar 29. PMID: 19329995
103. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018Jul;18(7):407-418 doi: 10.1038/s41568-018-0007-6. PMID: 29692415
104. Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and organoid model systems in cancer research. EMBO J. 2019Aug1;38(15):e101654 doi: 10.15252/embj.2019101654. Epub 2019 Jul 8. PMID: 31282586; PMCID: PMC6670015
105. Yang H, Wang Y, Wang P, Zhang N, Wang P. Tumor organoids for cancer research and personalized medicine. Cancer Biol Med. 2021Sep14;19(3):319-32 doi: 10.20892/j.issn.2095-3941.2021.0335. Epub ahead of print. PMID: 34520134; PMCID: PMC8958892
106. Lo YH, Kolahi KS, Du Y, Chang CY, Krokhotin A, Nair A, Sobba WD, Karlsson K, Jones SJ, Longacre TA, Mah AT, Tercan B, Sockell A, Xu H, Seoane JA, Chen J, Shmulevich I, Weissman JS, Curtis C, Califano A, Fu H, Crabtree GR, Kuo CJ. A CRISPR/Cas9-Engineered ARID1A-Deficient Human Gastric Cancer Organoid Model Reveals Essential and Nonessential Modes of Oncogenic Transformation. Cancer Discov. 2021Jun;11(6):1562-1581 doi: 10.1158/2159-8290.CD-20-1109. Epub 2021 Jan 15. PMID: 33451982; PMCID: PMC8346515
107. Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015Mar;21(3):256-62 doi: 10.1038/nm.3802. Epub 2015 Feb 23. PMID: 25706875
108. Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015May7;521(7550):43-7 doi: 10.1038/nature14415. Epub 2015 Apr 29. PMID: 25924068
109. Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, Matano M, Seino T, Nishikori S, Ishikawa K, Kawasaki K, Togasaki K, Takahashi S, Sukawa Y, Ishida H, Sugimoto S, Kawakubo H, Kim J, Kitagawa Y, Sekine S, Koo BK, Kanai T, Sato T. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell. 2018Aug9;174(4):856-869.e17 doi: 10.1016/j.cell.2018.07.027. PMID: 30096312
110. Smith RC, Tabar V. Constructing and Deconstructing Cancers using Human Pluripotent Stem Cells and Organoids. Cell Stem Cell. 2019Jan3;24(1):12-24 doi: 10.1016/j.stem.2018.11.012. Epub 2018 Dec 20. PMID: 30581078; PMCID: PMC6516073
111. Nam C, Ziman B, Sheth M, Zhao H, Lin DC. Genomic and Epigenomic Characterization of Tumor Organoid Models. Cancers (Basel). 2022Aug24;14(17):4090 doi: 10.3390/cancers14174090. PMID: 36077628; PMCID: PMC9454968
112. Guan X, Huang S. Advances in the application of 3D tumor models in precision oncology and drug screening. Front Bioeng Biotechnol. 2022Sep28;10:1021966 doi: 10.3389/fbioe.2022.1021966. PMID: 36246388; PMCID: PMC9555934
113. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015May7;161(4):933-45 doi: 10.1016/j.cell.2015.03.053. PMID: 25957691; PMCID: PMC6428276
114. Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017May;7(5):462-477 doi: 10.1158/2159-8290.CD-16-1154. Epub 2017 Mar 22. PMID: 28331002; PMCID: PMC5413423
115. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, van de Ven M, Egan DA, Zinzalla V, Moll J, Boj SF, Voest EE, Wessels L, van Diest PJ, Rottenberg S, Vries RGJ, Cuppen E, Clevers H. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018Jan11;172(1-2):373-386.e10 doi: 10.1016/j.cell.2017.11.010. Epub 2017 Dec 7. PMID: 29224780
116. de Witte CJ, Espejo Valle-Inclan J, Hami N, Lõhmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P, Nguyen L, Clevers H, Kloosterman WP, Cuppen E, Snippert HJG, Zweemer RP, Witteveen PO, Stelloo E. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020Jun16;31(11):107762 doi: 10.1016/j.celrep.2020.107762. PMID: 32553164
117. Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H, van Wijk LM, Revilla SA, Theeuwsen R, van de Ven M, van Roosmalen MJ, Ponsioen B, Ho VWH, Neel BG, Bosse T, Gaarenstroom KN, Vrieling H, Vreeswijk MPG, van Diest PJ, Witteveen PO, Jonges T, Bos JL, van Oudenaarden A, Zweemer RP, Snippert HJG, Kloosterman WP, Clevers H. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019May;25(5):838-849 doi: 10.1038/s41591-019-0422-6. Epub 2019 Apr 22. PMID: 31011202
118. Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, Bergren SK, Pietzak EJ, Anderson CB, Benson MC, Coleman JA, Taylor BS, Abate-Shen C, McKiernan JM, Al-Ahmadie H, Solit DB, Shen MM. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell. 2018Apr5;173(2):515-528.e17 doi: 10.1016/j.cell.2018.03.017. PMID: 29625057; PMCID: PMC5890941
119. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, IJzermans JNM, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017Dec;23(12):1424-1435 doi: 10.1038/nm.4438. Epub 2017 Nov 13. PMID: 29131160; PMCID: PMC5722201
120. Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, Chan D, Chan AS, Ma S, Lam KO, Bartfeld S, Man AHY, Lee BCH, Chan ASY, Wong JWH, Cheng PSW, Chan AKW, Zhang J, Shi J, Fan X, Kwong DLW, Mak TW, Yuen ST, Clevers H, Leung SY. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell. 2018Dec6;23(6):882-897.e11 doi: 10.1016/j.stem.2018.09.016. Epub 2018 Oct 18. PMID: 30344100
121. Tao Y, Kang B, Petkovich DA, Bhandari YR, In J, Stein-O'Brien G, Kong X, Xie W, Zachos N, Maegawa S, Vaidya H, Brown S, Chiu Yen RW, Shao X, Thakor J, Lu Z, Cai Y, Zhang Y, Mallona I, Peinado MA, Zahnow CA, Ahuja N, Fertig E, Issa JP, Baylin SB, Easwaran H. Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and BrafV600E-Induced Tumorigenesis. Cancer Cell. 2019Feb11;35(2):315-328.e6 doi: 10.1016/j.ccell.2019.01.005. PMID: 30753828; PMCID: PMC6636642
122. Nakaoka T, Saito Y, Shimamoto Y, Muramatsu T, Kimura M, Kanai Y, Saito H. Cluster microRNAs miR-194 and miR-215 suppress the tumorigenicity of intestinal tumor organoids. Cancer Sci. 2017Apr;108(4):678-684 doi: 10.1111/cas.13165. Epub 2017 Apr 19. PMID: 28092415; PMCID: PMC5406536
123. Bartfeld S, Clevers H. Organoids as Model for Infectious Diseases: Culture of Human and Murine Stomach Organoids and Microinjection of Helicobacter Pylori. J Vis Exp. 2015Nov12(105):53359 doi: 10.3791/53359. PMID: 26650279; PMCID: PMC4692704
124. Rao S, Hossain T, Mahmoudi T. 3D human liver organoids: An in vitro platform to investigate HBV infection, replication and liver tumorigenesis. Cancer Lett. 2021May28;506:35-44 doi: 10.1016/j.canlet.2021.02.024. Epub 2021 Mar 4. PMID: 33675983
125. Toyohara Y, Taguchi A, Ishii Y, Yoshimoto D, Yamazaki M, Matsunaga H, Nakatani K, Hoshi D, Tsuchimochi S, Kusakabe M, Baba S, Kawata A, Ikemura M, Tanikawa M, Sone K, Uchino-Mori M, Ushiku T, Takeyama H, Oda K, Kawana K, Hippo Y, Osuga Y. Identification of target cells of human papillomavirus 18 using squamocolumnar junction organoids. Cancer Sci. 2024Jan;115(1):125-138 doi: 10.1111/cas.15988. Epub 2023 Nov 23. PMID: 37996972; PMCID: PMC10823277
126. Veninga V, Voest EE. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell. 2021Sep13;39(9):1190-1201 doi: 10.1016/j.ccell.2021.07.020. Epub 2021 Aug 19. PMID: 34416168
127. Naruse M, Ochiai M, Sekine S, Taniguchi H, Yoshida T, Ichikawa H, Sakamoto H, Kubo T, Matsumoto K, Ochiai A, Imai T. Re-expression of REG family and DUOXs genes in CRC organoids by co-culturing with CAFs. Sci Rep. 2021 Jan 22;11(1):2077. doi: 10.1038/s41598-021-81475-2. Erratum in: Sci Rep. 2021Jul29;11(1):15801 PMID: 33483567; PMCID: PMC7822883
128. Liu C, Yang M, Zhang D, Chen M, Zhu D. Clinical cancer immunotherapy: Current progress and prospects. Front Immunol. 2022Oct11;13:961805 doi: 10.3389/fimmu.2022.961805. PMID: 36304470; PMCID: PMC9592930
129. Sun CP, Lan HR, Fang XL, Yang XY, Jin KT. Organoid Models for Precision Cancer Immunotherapy. Front Immunol. 2022Apr5;13:770465 doi: 10.3389/fimmu.2022.770465. PMID: 35450073; PMCID: PMC9016193
130. Marrella A, Varani G, Aiello M, Vaccari I, Vitale C, Mojzisek M, Degrassi C, Scaglione S. 3D fluid-dynamic ovarian cancer model resembling systemic drug administration for efficacy assay. ALTEX. 2021;38(1):82-94 doi: 10.14573/altex.2003131. Epub 2020 Aug 3. PMID: 32754773. (a)
131. Marrella A, Fedi A, Varani G, Vaccari I, Fato M, Firpo G, Guida P, Aceto N, Scaglione S. High blood flow shear stress values are associated with circulating tumor cells cluster disaggregation in a multi-channel microfluidic device. PLoS One. 2021Jan14;16(1):e0245536 doi: 10.1371/journal.pone.0245536. PMID: 33444361; PMCID: PMC7808575. (b)
132. Marrella A, Buratti P, Markus J, Firpo G, Pesenti M, Landry T, Ayehunie S, Scaglione S, Kandarova H, Aiello M. In vitro demonstration of intestinal absorption mechanisms of different sugars using 3D organotypic tissues in a fluidic device. ALTEX. 2020;37(2):255-264 doi: 10.14573/altex.1908311. Epub 2019 Dec 30. PMID: 31893489
133. Fedi A, Vitale C, Ponschin G, Ayehunie S, Fato M, Scaglione S. In vitro models replicating the human intestinal epithelium for absorption and metabolism studies: A systematic review. J Control Release. 2021Jul10;335:247-268 doi: 10.1016/j.jconrel.2021.05.028. Epub 2021 May 24. PMID: 34033859
134. Schimmel L, Heemskerk N, van Buul JD. Leukocyte transendothelial migration: A local affair. Small GTPases. 2017Jan2;8(1):1-15 doi: 10.1080/21541248.2016.1197872. Epub 2016 Aug 15. PMID: 27715453; PMCID: PMC5331897
135. Huang Q, Hu X, He W, Zhao Y, Hao S, Wu Q, Li S, Zhang S, Shi M. Fluid shear stress and tumor metastasis. Am J Cancer Res. 2018May1;8(5):763-777 PMID: 29888101; PMCID: PMC5992512
136. Lapizco-Encinas BH, Zhang YV. Microfluidic systems in clinical diagnosis. Electrophoresis. 2023Jan;44(1-2):217-245 doi: 10.1002/elps.202200150. Epub 2022 Sep 3. PMID: 35977346
137. Ko J, Park D, Lee S, Gumuscu B, Jeon NL. Engineering Organ-on-a-Chip to Accelerate Translational Research. Micromachines (Basel). 2022Jul28;13(8):1200 doi: 10.3390/mi13081200. PMID: 36014122; PMCID: PMC9412404
138. Pulsoni I, Lubda M, Aiello M, Fedi A, Marzagalli M, von Hagen J, Scaglione S. Comparison Between Franz Diffusion Cell and a novel Micro-physiological System for In Vitro Penetration Assay Using Different Skin Models. SLAS Technol. 2022Jun;27(3):161-171 doi: 10.1016/j.slast.2021.12.006. Epub 2022 Jan 2. PMID: 35058208
139. Sun W, Luo Z, Lee J, Kim HJ, Lee K, Tebon P, Feng Y, Dokmeci MR, Sengupta S, Khademhosseini A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv Healthc Mater. 2019 Feb;8(4):e1801363. doi: 10.1002/adhm.201801363. Epub 2019 Jan 3. Erratum in: Adv Healthc Mater. 2019Aug;8(15):e1900754 PMID: 30605261; PMCID: PMC6424124
140. Trujillo-de Santiago G, Flores-Garza BG, Tavares-Negrete JA, Lara-Mayorga IM, González-Gamboa I, Zhang YS, Rojas-Martínez A, Ortiz-López R, Álvarez MM. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials (Basel). 2019Sep11;12(18):2945 doi: 10.3390/ma12182945. PMID: 31514390; PMCID: PMC6766252
141. Marzagalli M, Pelizzoni G, Fedi A, Vitale C, Fontana F, Bruno S, Poggi A, Dondero A, Aiello M, Castriconi R, Bottino C, Scaglione S. A multi-organ-on-chip to recapitulate the infiltration and the cytotoxic activity of circulating NK cells in 3D matrix-based tumor model. Front Bioeng Biotechnol. 2022Jul25;10:945149 doi: 10.3389/fbioe.2022.945149. PMID: 35957642; PMCID: PMC9358021
142. Lim LS, Hu M, Huang MC, Cheong WC, Gan AT, Looi XL, Leong SM, Koay ES, Li MH. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab Chip. 2012Nov7;12(21):4388-96 doi: 10.1039/c2lc20750h. PMID: 22930096
143. Wang S, Wan Y, Liu Y. Effects of nanopillar array diameter and spacing on cancer cell capture and cell behaviors. Nanoscale. 2014Nov7;6(21):12482-9 doi: 10.1039/c4nr02854f. PMID: 25137436; PMCID: PMC4194151
144. Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004Jan;35(1):122-8 doi: 10.1016/j.humpath.2003.08.026. PMID: 14745734
145. Kimura H, Sakai Y, Fujii T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab Pharmacokinet. 2018Feb;33(1):43-48 doi: 10.1016/j.dmpk.2017.11.003. Epub 2017 Nov 13. PMID: 29175062
146. Yan J, Li Z, Guo J, Liu S, Guo J. Organ-on-a-chip: A new tool for in vitro research. Biosens Bioelectron. 2022Nov15;216:114626 doi: 10.1016/j.bios.2022.114626. Epub 2022 Aug 10. PMID: 35969963
147. Fedi A, Vitale C, Fato M, Scaglione S. A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays. Bioengineering (Basel). 2023Feb18;10(2):270 doi: 10.3390/bioengineering10020270. PMID: 36829764; PMCID: PMC9952600
148. Guo QR, Zhang LL, Liu JF, Li Z, Li JJ, Zhou WM, Wang H, Li JQ, Liu DY, Yu XY, Zhang JY. Multifunctional microfluidic chip for cancer diagnosis and treatment. Nanotheranostics. 2021Jan1;5(1):73-89 doi: 10.7150/ntno.49614. PMID: 33391976; PMCID: PMC7738943
149. Vanderburgh J, Sterling JA, Guelcher SA. 3D Printing of Tissue Engineered Constructs for In Vitro Modeling of Disease Progression and Drug Screening. Ann Biomed Eng. 2017Jan;45(1):164-179 doi: 10.1007/s10439-016-1640-4. Epub 2016 May 11. PMID: 27169894; PMCID: PMC5106334
150. Groll J, Burdick JA, Cho DW, Derby B, Gelinsky M, Heilshorn SC, Jüngst T, Malda J, Mironov VA, Nakayama K, Ovsianikov A, Sun W, Takeuchi S, Yoo JJ, Woodfield TBF. A definition of bioinks and their distinction from biomaterial inks. Biofabrication. 2018Nov23;11(1):013001 doi: 10.1088/1758-5090/aaec52. PMID: 30468151
151. Heid S, Boccaccini AR. Advancing bioinks for 3D bioprinting using reactive fillers: A review. Acta Biomater. 2020Sep1;113:1-22 doi: 10.1016/j.actbio.2020.06.040. Epub 2020 Jul 2. PMID: 32622053
152. Stanton MM, Samitier J, Sánchez S. Bioprinting of 3D hydrogels. Lab Chip. 2015Aug7;15(15):3111-5 doi: 10.1039/c5lc90069g. PMID: 26066320
153. Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014Aug;32(8):773-85 doi: 10.1038/nbt.2958. PMID: 25093879
154. Han S, Kim S, Chen Z, Shin HK, Lee SY, Moon HE, Paek SH, Park S. 3D Bioprinted Vascularized Tumour for Drug Testing. Int J Mol Sci. 2020Apr23;21(8):2993 doi: 10.3390/ijms21082993. PMID: 32340319; PMCID: PMC7215771
155. Jung M, Skhinas JN, Du EY, Tolentino MAK, Utama RH, Engel M, Volkerling A, Sexton A, O'Mahony AP, Ribeiro JCC, Gooding JJ, Kavallaris M. A high-throughput 3D bioprinted cancer cell migration and invasion model with versatile and broad biological applicability. Biomater Sci. 2022Oct11;10(20):5876-5887 doi: 10.1039/d2bm00651k. PMID: 36149407
156. Robin Augustine, Sumama Nuthana Kalva, Rashid Ahmad, Alap Ali Zahid, Shajia Hasan, Ajisha Nayeem, Lana McClements, Anwarul Hasan. 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Translational Oncology. 2021;4:101015 101015, ISSN 1936-5233, https://doi.org/10.1016/j.tranon.2021.101015
157. Charbe N, McCarron PA, Tambuwala MM. Three-dimensional bio-printing: A new frontier in oncology research. World J Clin Oncol. 2017Feb10;8(1):21-36 doi: 10.5306/wjco.v8.i1.21. PMID: 28246583; PMCID: PMC5309712
158. Sánchez-Salazar MG, Crespo-López Oliver R, Ramos-Meizoso S, Jerezano-Flores VS, Gallegos-Martínez S, Bolívar-Monsalve EJ, Ceballos-González CF, Trujillo-de Santiago G, Álvarez MM. 3D-Printed Tumor-on-Chip for the Culture of Colorectal Cancer Microspheres: Mass Transport Characterization and Anti-Cancer Drug Assays. Bioengineering (Basel). 2023May5;10(5):554 doi: 10.3390/bioengineering10050554. PMID: 37237624; PMCID: PMC10215397
Author contact
![]() Corresponding authors: Ciro Isidoro, D.Sc., M.D., e-mail: ciro.isidorouniupo.it, Tel.: +39-0321-660507; Fax: +39-0321-620421; Alessandra Ferraresi, Ph.D., e-mail: alessandra.ferraresiuniupo.it.
Corresponding authors: Ciro Isidoro, D.Sc., M.D., e-mail: ciro.isidorouniupo.it, Tel.: +39-0321-660507; Fax: +39-0321-620421; Alessandra Ferraresi, Ph.D., e-mail: alessandra.ferraresiuniupo.it.

 Global reach, higher impact
Global reach, higher impact