10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(1):63-74. doi:10.7150/ijbs.101861 This issue Cite
Review
Targeting protein modification: a new direction for immunotherapy of pancreatic cancer
1. Department of Hepatobiliary and Pancreatic Surgery, Northern Jiangsu People's Hospital Affiliated to Yangzhou University, Jiangsu 225000, China.
2. The Yangzhou School of Clinical Medicine of Dalian Medical University, Jiangsu 225000, China.
† These authors contributed equally to this research.
Received 2024-8-3; Accepted 2024-11-3; Published 2025-1-1
Abstract
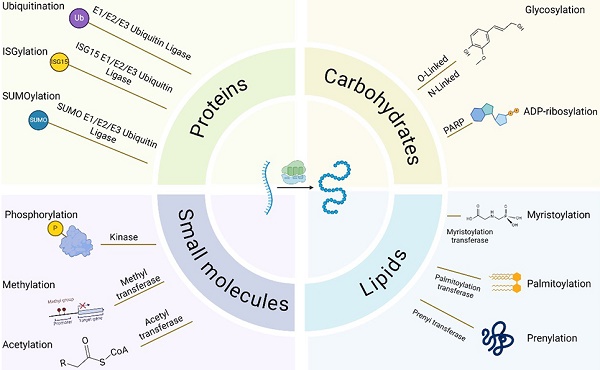
Post-translational modifications (PTMs) alter protein conformation by covalently attaching functional groups to substrates, influencing their biological activity, mechanisms of action, and functional performance. PTMs and their interactions are essential to many critical signal transduction processes, including tumor transformation, cancer progression, and metastasis in pancreatic cancer. Additionally, advancements in tumor immunotherapy indicate that PTMs are essential in immune cell activation, transport, and energy metabolism. This study aimed to investigate the effects of different PTMs on immunotherapy for pancreatic cancer, providing new perspectives and suggesting directions for future research.
Keywords: Post-translational modifications, Immunotherapy, Targeted therapy, Immune escape, Pancreatic cancer
Introduction
Pancreatic cancer predominantly comprises pancreatic ductal adenocarcinoma (PDAC), a malignancy associated with poor prognosis [1]. In 2022, approximately 510,566 new cancer cases were reported globally, resulting in 467,005 deaths, which makes it the sixth most common cause of cancer-related deaths globally [2]. The 5-year survival rate for patients with stage I or II PDAC after surgery is 24.6% [3]. Despite the advancements in treatments, including chemotherapy, neoadjuvant, and targeted therapies, significant challenges persist [4].
Immunotherapy is a new and effective treatment option [5]. It primarily includes immune checkpoint inhibitors (ICIs), adoptive cell therapy (ACT), and monoclonal antibody therapy [6]. ACT is a highly personalized treatment that targets cancer through the transplantation of autologous or allogeneic tumor-specific T cells [7]. Significant advancements have been made in adoptive cell therapies, including Chimeric Antigen Receptor T-Cell Immunotherapy (CAR-T) and T-cell receptor-engineered T cell (TCR-T) cell therapies [8, 9]. Monoclonal antibody is a type of targeted therapy characterized by significant specificity, extended serum half-life, strong binding affinity, and ability to activate immune effector functions [10].
Despite significant advancements in immunotherapy, the near-universal resistance of PDAC to immunotherapy is a significant exception in human cancers. Effective responses are observed in less than 1% of patients with microsatellite instability-high (MSI-H) tumors. The tumor microenvironment (TME) of PDAC is frequently described as "cold," marked by the limited presence of effector T cells and a significant influx of myeloid cells [11, 12]. Furthermore, features including a low mutational burden and an immunosuppressive TME hinder T cell activation, migration, and functionality, thereby exacerbating the challenges to adaptive immunity in PDAC [13, 14]. The most promising strategy for PDAC treatment entails comprehensive study and optimization of immunotherapy, shifting focus from solely targeting malignant cell proliferation and invasion to investigating the complex interactions between tumors and TME.
Post-translational modifications (PTMs) influence the complexity and diversity of the proteome by covalently binding functional groups to substrate proteins. The method involves adding various functional groups into the side chains of amino acids, including acetyl, phosphate, sugar, and methyl groups. The polypeptide chain undergoes multiple PTMs within different cellular compartments, including the nucleus, cytosol, endoplasmic reticulum, and Golgi apparatus [15]. During physiologic and pathologic conditions, it can enhance the functional diversity of proteins by modulating protein folding, activity, stability, localization, signal transduction, and binding [16, 17]. Consequently, these modifications are essential in various physiological activities, including signal transduction, gene expression, and cell cycle regulation [18, 19]. With the enhanced accessibility of genomic sequencing data and the rapid development of detection methods, over 600 types of PTMs have been discovered to date [20]. The most common include protein phosphorylation, acetylation, SUMOylation, glycosylation, and palmitoylation (Fig. 1).
PTMs are essential in immune activity in the body, significantly influencing immune cell activation, signal regulation, immune response, and tumor metabolic reprogramming. They regulate TME by affecting immune cell differentiation and function [21-25]. PTMs can directly or indirectly affect the efficacy of immunotherapy by regulating immune checkpoints or altering the TME [26]. PTMs can regulate the immunogenic characteristics of cancer cells, affecting their recognition and susceptibility to immune system attacks [24].
Because protein PTMs regulate cancer development and progression, examining these alterations in the context of immune responses may offer a comprehensive understanding of the mechanisms regulating interactions between cancer cells and immune cells. Herein, we systematically examine and present the recent advancements regarding the role of PTMs in the immunotherapy of PDAC.
Phosphorylation
Phosphorylation is a classic and reversible PTM prevalent in eukaryotes. In mammals, approximately 30% of proteins undergo phosphorylation, a process dynamically regulated by protein kinases and phosphatases [27]. This alteration is essential for numerous cellular activities, including cell division, membrane transport, gene expression regulation, and protein interactions [28]. Numerous phosphorylation events have been identified in PDAC (Table 1). Furthermore, phosphorylation is essential in tumor immunotherapy.
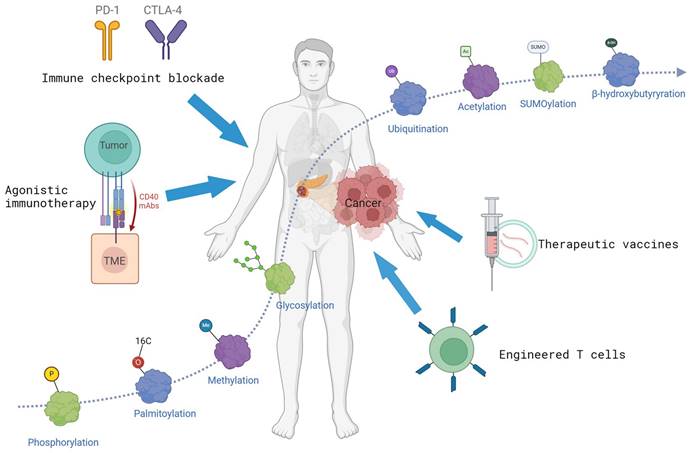
PTMs in immunotherapy of pancreatic cancer. Many proteins and PTMs (such as phosphorylation, acetylation, ubiquitination, etc.) are implicated in tumorigenesis. PTMs can influence the efficacy of immunotherapy. The figure is generated with BioRender (https://biorender.com).
Identifying phosphorylation targets associated with PDAC
| Target | Function in cancer | Reference |
|---|---|---|
| FAM83A | Promote the transcriptional activity of β-catenin | [141] |
| PDE4D | Control the degradation of Camp. | [142] |
| Girdin | Control the cytoskeleton and vascular remodeling | [143] |
| ASPP2 | Regulating cell apoptosis. | [144] |
| WAVE3 | Promote epithelial mesenchymal transition and regulate metastasis. | [145] |
| IER3 | Activate ERK1/2 to support the development of PanIN after pancreatitis. | [146] |
| MUC4 | The transmembrane ligand of ERBB2 maintains its stability on the plasma membrane and enhances activation. | [147] |
| Stattic | Inhibition of STAT3 activation and nuclear translocation. | [148] |
| CAP1 | Regulating actin cytoskeleton and cell migration. | [149] |
| CTDSPL2 | Regulating mitosis and promoting cell movement. | [150] |
| IQGAP1 | articipate in cytoskeleton remodeling, cell migration and intercellular signal transduction. | [151] |
A primary challenge in tumor immunotherapy research is investigating the immune evasion mechanisms associated with immune checkpoints. According to their targets, immune checkpoint inhibitors include Programmed Cell Death Protein 1 (PD-1) inhibitors, Programmed Death-Ligand 1 (PD-L1) inhibitors, and Cytotoxic T-lymphocyte associated protein 4 (CTLA-4) inhibitors. The interaction between PD-L1 and PD-1 exerts a negative regulatory effect, facilitating peripheral immune tolerance [29]. The regulatory role of the PDCD1 gene is well-established [30]; however, PTMs are significant factors affecting PD-1 and PD-L1 interaction. Zhang et al. [31] reported that the limited efficacy of immunotherapy in PDAC is primarily due to PD-L1 dephosphorylation by Never in Mitosis A-related kinase 2 (NEK2). NEK2 enhances PD-L1 stability by inhibiting its proteasomal degradation through phosphorylation at T194/T210 residues and further stabilizes PD-L1 through glycosylation at N192, N200, and N219 sites [31, 32].
Gemcitabine (GEM), the most commonly used chemotherapeutic agent for PDAC, induces Signal Transducer and Activator of Transcription 1 (STAT1) phosphorylation after treatment and elicits various PD-L1-inducing cytokines, including IFN-γ, IL-6, and TNF-α [33]. A previous study reported that statins combined with GEM inhibit STAT1 phosphorylation, significantly reducing PD-L1 expression and enhancing CD8+ T cell infiltration [34]. Moreover, statins reduce YAP/TAZ expression through AKT phosphorylation, further inhibiting PD-L1 expression [35]. In TME, neutrophil extracellular traps induce T cell exhaustion and dysfunction via PD-L1, a mechanism closely linked to the chemotactic effect of CXC motif chemokine receptor 2 (CXCR2) on neutrophils [36-38], a mechanism closely linked to the chemotactic effect of CXCR2 on neutrophils, inhibits the recruitment and function of CXCR2 in neutrophils by inducing STAT1 dephosphorylation at Tyr701 in these cells. In animal models, Nifurtimox significantly enhances PDAC sensitivity to GEM and PD-1 blockade therapy [39, 40]. Zhang et al. [41] reported that Polo-like Kinase 1 (Plk1) induces retinoblastoma protein (RB) phosphorylation at S758, leading to dysregulated NF-κB translocation and increased PD-L1 expression. Inhibition of Plk1 enhances sensitivity to immune checkpoint blockade. NSG3, a vesicular transport protein, is a potential diagnostic and prognostic marker that inhibits PDAC cell proliferation and invasion and suppresses Erk1/2 phosphorylation, thereby inhibiting PD-L1 expression and improving immunotherapy outcomes [42, 43].
T cells are essential in protecting the body from pathogens. Enhancing T-cell infiltration can significantly improve the efficacy of existing cancer immunotherapies, including ICB therapy [44, 45]. T-cell activity is regulated by the phosphorylation of specific proteins or enzymes within the tumor. In patients with PDAC, the inhibition of IRAK4 phosphorylation in tumor cells downregulates Hyaluronan synthase 2 (HAS2) through an NF-κB-dependent mechanism. This reduction in HAS2 levels mitigates T-cell exhaustion and enhances responsiveness to checkpoint immunotherapies, including anti-CTLA-4 and anti-PD-1 [46, 47]. The T cell receptor (TCR) is activated by phosphorylation at Tyr-323 (pY323), which binds to p38-activated MAPK as an alternative pathway for p38 activation. TCR-mediated activation of CD4+ tumor-infiltrating lymphocytes (TILs) leads to alternative p38 activation and pro-inflammatory cytokine production [48]. Targeting this alternative p38 pathway in T cells demonstrates promising preventive and therapeutic effects in PDAC models by disrupting downstream pro-inflammatory pathways [49]. Interleukin-35 (IL-35), a cytokine of the IL-12 family primarily produced by CD4+ T cells and B cells, induces STAT3 phosphorylation. This process inhibits CD8+ T-cell infiltration and activation and promotes tumor growth [50, 51]. Targeting IL-35 to enhance T-cell infiltration and transform the TME of PDAC from "cold" to "hot" is an effective strategy to improve the efficacy of immunotherapy. Chemotherapy should address immune suppression within tumors to achieve adequate therapeutic outcomes. PX-478, for example, can inhibit HIF-1α expression, increase eIF2α phosphorylation levels, enhance GEM immunogenicity, and strengthen cytotoxic T-cell responses against PDAC cells [52].
The immune response in PDAC is partially regulated by immunosuppressive myeloid cells, rendering these cells a promising target for immunotherapy [53]. Macrophages are among the most abundant immune cell types in the TME and facilitate tumor progression by creating an immunosuppressive TME from the early stages [54, 55]. Re-polarizing tumor-associated macrophages (TAMs) toward an M1-like phenotype has been proposed as a potential therapeutic option for cancer [56]. Dual-specificity tyrosine-regulated kinase 1B (DYRK1B), a kinase that regulates tyrosine phosphorylation, is present in 90% of pancreatic cancer cases and is negatively correlated with macrophages in tumor tissues [57, 58]. Inhibiting DYRK1B in TAMs accelerates their polarization toward an M1 phenotype, thereby reducing cancer cell surface marker CD24 expression [59]. This, subsequently, enhances immune cell identification and eradication of cancer cells [58]. STAT3 is essential in solid tumor progression [60, 61]. Phosphorylated STAT3, as a direct target of miR-506, reprograms M2-polarized macrophages into an M1 phenotype, thereby reversing the immunosuppressive microenvironment in PDAC.
Natural killer (NK) cells are innate lymphocytes, and their activation is regulated by the interactions between NK receptors and target cells [62, 63]. This makes NK cell-based therapies a significant focus of innovation in immunotherapy. Two NK cell subsets can be identified in human peripheral blood: (1) the CD56bright subset, which secretes immunoregulatory cytokines, and (2) the CD56dim subset, comprising approximately 90% of the total number of NK cells, which exerts cytotoxicity through the cell surface Fc receptor CD16 [64].
The NF-κB signaling pathway regulates the differentiation of NK cell subsets and their immune responses. Phosphorylation of iκB protein in PDAC cells facilitates nuclear translocation of NF-κB. The activation of NF-κB subsequently promotes CXCL8 and the transcription factor P65 transcription, facilitating the migration of radiation-induced CD56dim NK cells from tumor cells and inhibiting tumor growth. This indicates that combining NK cell adoptive therapy with radiotherapy can effectively induce tumor cell apoptosis [65]. A previous study has reported that polysaccharides enhance the antitumor effects of GEM through the activation of NK cells [66]. In PDAC, polysaccharides secreted by SEP bind to the TLR4 receptor on NK cells, upregulating ERK, JNK, p38, and NF-κB phosphorylation levels. TLR4/MAPKs/NF-κB signaling pathway activation increases NKG2D expression in NK cells, thereby synergistically enhancing the anti-pancreatic tumor effects of GEM [67]. In addition, NK cells can be combined with therapeutic antibodies for cancer treatment [68]. Enhancing NK cell FcR effector functions through Interleukin-21 (IL-21) is a promising strategy to improve the efficacy of cetuximab. Following IL-21 interaction with NK cell surface receptors, the STAT1 phosphorylation level increases. When NK cells are stimulated by cetuximab-coated tumor cells, the ERK phosphorylation level increases, leading to intracellular activation of NK cells, reduced tumor burden, and improved therapeutic outcomes [69].
Glycosylation
Glycosylation is essential for stabilizing membrane protein expression and preserving normal physiological function [70]. Eight glycosylation pathways have been identified, with N-glycosylation and O-glycosylation significantly associated with disease progression. PDAC tumors demonstrate distinct alterations in glycosylation (Table 2), including an increased abundance of the sialic acid Lewis A antigen CA19-9 [71]. Besides, PDACs have elevated levels of fucosylated and branched and truncated O-glycans [72-74], which are associated with tumor progression and poor prognosis (Table 2). Additionally, abnormal glycosylation contributes to tumor immune evasion [75]. Consequently, glycosylation is a potential target for anticancer therapy [76].
Glycosylation targets associated with PDAC
| Target | Function in cancer | Reference |
|---|---|---|
| ST3Gal1 | Attaching sialic acid to T-antigen, producing sialyl T-antigen | [152] |
| MUC1 | Activating the EGFR-PI3K/Akt signaling pathway and help cancer cells fight anoikis | [153] |
| CA199 | Pancreatic cancer tumor biomarkers | [154] |
| MDH1 | Involved in the interconversion of pyruvate and malic acid in mitochondria | [155] |
| CD44 | Promoting the expression of NANOG in pancreatic cancer cells and facilitate the alteration of CSC feature | [156] |
| MGAT5 | N-glycan branching through adding β1,6-linked N-acetylglucosamine (β1,6-GlcNAc) to an α1,6-linked mannose | [86] |
| TNFR1 | Increased α-2,6-sialylation of TNFR1 inhibits internalization and stabilizes signaling through AKT and NF-κB, conferring resistance to gemcitabine and TNF-induced apoptosis | [157] |
Sialylation is a pervasive and complex form of glycosylation that has become a target for cancer therapy because of its immunosuppressive properties [77]. In PDAC, the elevated ST3Gal1 and ST3Gal14 expression results in increased α2,3-sialylation on tumor cells, facilitating the differentiation of monocytes into immunosuppressive TAMs by binding to myeloid cell receptors Siglec-7 and -9 [75]. Furthermore, excessive tumor sialylation inhibits NK cell activity and disrupts Teff/Treg balance, facilitating immune escape [78]. Salivation inhibition enhances the immune response through various mechanisms, including facilitating dendritic cell maturation, increasing the number and activation status of effector immune cells, particularly CD8+ T cells, and enhancing cytotoxic T cell activities [79].
Mucin is an essential defense barrier in the body that is frequently overexpressed and abnormally glycosylated in PDAC, acting as a source of tumor-associated antigens and potential therapeutic targets. Mucin 4 (MUC4) is a compelling TAA. Wei et al. [80] first reported that transducing dendritic cells with pan-DR helper T cell epitopes or the universal T epitope (PADRE) with HLA-A1 and HLA-A2-specific MUC4 epitopes led to the upregulation of dendritic cell activation markers including HLA-DR, CD80, and CD86, thereby inducing an effective MUC4-specific cytotoxic T cell response. Furthermore, glycopeptide immunization using glycosylated MUC4 tandem repeat peptides has demonstrated effective antigen-specific immune responses in PDAC mouse models [81]. Mucin 1 (MUC1) is a protein composed of repetitive 20-amino-acid sequences and undergoes extensive O-glycosylation. In PDAC, MUC1 glycosylation is irregularly distributed, which enhances the recognition of the immune system and processing of the protein structure, consequently eliciting an immune response against MUC1 [81]. A phase I/II clinical trial demonstrated that dendritic cell vaccines infused with MUC1 significantly enhanced CD8 and CD4 T cell activities, effectively improving immunosuppression in patients undergoing pancreatic surgery [82].
Engineered T cells expressing chimeric antigen receptors (CARs) signify a promising research focus in immunotherapy [83]. However, CAR T therapy encounters challenges, including inefficient delivery and penetration to tumor sites, and its efficacy depends on the density and accessibility of tumor cell antigens [84]. Abnormal glycosylation in tumor cells manifests as an extracellular glycan layer on the cell surface. This glycoprotein shell can participate in basic biological processes and disrupt immune responses by masking immune cell epitopes. Glycosyltransferase 5 (MGAT5) is an essential gene that regulates N-glycan chains [85]. MGAT5-derived N-glycans provide strong protection against pancreatic cancer [86]. In PDAC, defects in N-glycosylation due to MGAT5 knockout induce robust immune synapses between tumor cells and 44v6 CAR T cells. This interaction is characterized by increased F-actin accumulation, enhanced granule convergence, and a reduced distance from the microtubule organizing center to F-actin. In PDAC with N-glycosylation defects, activated 44v6 T cells exhibit enhanced signaling of the calcium-dependent phosphatase nuclear factor and NF-κB, thereby enhancing CAR T cell efficacy in the immune response against PDAC [87]. On the other hand, tumor polysaccharide coating can also be used as a marker for immunotherapy [88]. For instance, 5E5 CAR T cells specifically target the Tn-MUC1 glycopeptide epitope on PDAC cell surfaces, resulting in significant tumor accumulation and exhibiting significant anticancer efficacy in mouse models [89].
Acetylation
Acetylation is a dynamic and reversible PTM in which acetyl groups are transferred to substrates by acetyltransferases. However, proteins undergo deacetylation through the action of deacetylases [90] (Table 3). This process, termed histone acetylation, was initially identified in histones [91]. In mammals, acetylation occurs on non-histone lysine residues, including those in high mobility group proteins, tubulin, and p53 [92]. Consequently, the enzymes involved are reclassified as lysine acetyltransferases [93]. Acetylation has been implicated in the pathogenic processes of pancreatic cancer development (Table 3). Recent studies highlight the importance of acetylation modifications in immune system function and tumor immunity [94].
Identifying acetylation targets associated with PDAC
| Target | Function in cancer | Reference |
|---|---|---|
| P65 | Deacetylation at the P65 K310 site inhibits NF-κB transcriptional activity and inhibits PD-L1 expression | [97] |
| SIRT5 | SIRT5 loss enhanced glutamine and glutathione metabolism via acetylation-mediated activation of GOT1 | [158] |
| HSPA5 | Acetylation at K353 site of HSPA5 promoted ferroptosis of PDAC | [159] |
| BCAT2 | BCAT2 acetylation suppresses BCAA catabolism and pancreatic tumor growth | [160] |
| PGC-1α | PGC-1α acetylation causes metabolism to shift from a mitochondrial oxidative catabolic process to fatty acid synthesis | [161] |
| STAT3 | STAT3 acetylation inhibits the STAT3/SIRT1 interaction and enhances the function of immunosuppressive cells in pancreatic cancer | [162] |
Histone deacetylases (HDAC) can remove acetyl groups from acetylated proteins [95]. HDACs have garnered attention for their role in immune evasion, making them a promising target for therapeutic strategies [96]. Currently, there are five HDAC inhibitors approved for clinical use. These inhibitors can disrupt PD-L1 and PD-1 interaction. HDAC5 inhibits immune responses and increases T regulatory cells, highlighting its significance in antitumor immunity. In PDAC, inhibition of HDAC5 suppresses NF-κB-mediated PD-L1 expression and improves the efficacy of anti-PD-1 therapy. Therefore, HDAC inhibitors can enhance the sensitivity of PDAC to immune checkpoint therapy [97]. The HDAC inhibitor LBH589 can enhance histone acetylation in the PD-L1 promoter region, thereby rapidly enhancing PD-L1 expression [98]. Moreover, HDAC3 inhibitors, including RGFP966, reduce PD-L1 mRNA and protein levels, thereby enhancing immune surveillance and reversing immune evasion [99]. Chin-King Looi et al. [100] found that HDAC inhibitors givinostat and dacinosta can reverse the sensitivity of Cytotoxic T lymphocytes (CTLs) resistant PDAC cells to CTLs. Furthermore, HDAC inhibitors can mitigate immune evasion by reprogramming tumor-associated myeloid-derived suppressor cells (MDSCs). Entinostat reprograms MDSCs in pancreatic tumor models, transforming immune-resistant tumors into those responsive to checkpoint therapies [101].
Ubiquitination
Ubiquitination is the binding of ubiquitin to specific amino acids as monomers or polymers. Ubiquitin-activating enzymes facilitate this process and depend on the synchronized function of three essential proteins: the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3) [102]. E3 ligases are essential for recognizing specific substrate proteins, thereby tightly regulating ubiquitination. Deubiquitinating enzymes (DUBs) reverse the process. Emerging evidence indicates that ubiquitination and deubiquitination play key roles in regulating the progression and prognosis of pancreatic cancer (Table 4). The dynamic equilibrium between ubiquitination and deubiquitination regulates protein expression levels, ensuring protein function stability, which ultimately affects substrate activity [103, 104].
Identifying ubiquitination and deubiquitination targets associated with PDAC
| Target | Function in cancer | Reference |
|---|---|---|
| USP8 | USP8 interacts with PD-L1 to inhibit its ubiquitination proteasome degradation | [111] |
| USP10 | USP10 inhibits YAP1 ubiquitination and degradation to promote Cyr61 expression, which induces immune escape and promotes growth and metastasis of PAAD | [109] |
| USP25 | USP25 regulates HIF-1α transcriptional activity and regulates metabolic reprogramming, promoting PDAC cell growth | [163] |
| USP22 | USP22 deubiquitinated PD-L1 and inhibited its proteasome degradation | [164] |
| cGAS | The ubiquitination degradation of cGAS inhibited the activation of CGAS-STING signaling pathway and reduced the production of pro-inflammatory cytokines and type I interferon | [165] |
| β-catenin | Ubiquitination degradation of β-catenin leads to cell cycle arrest at G1 and promotes apoptosis | [166] |
| eEF1A1 | eEF1A1 acts with FBXO32 to promote ubiquitination of eEF1A1 at K273, enhancing its activity and increasing protein synthesis in PDAC cells | [167] |
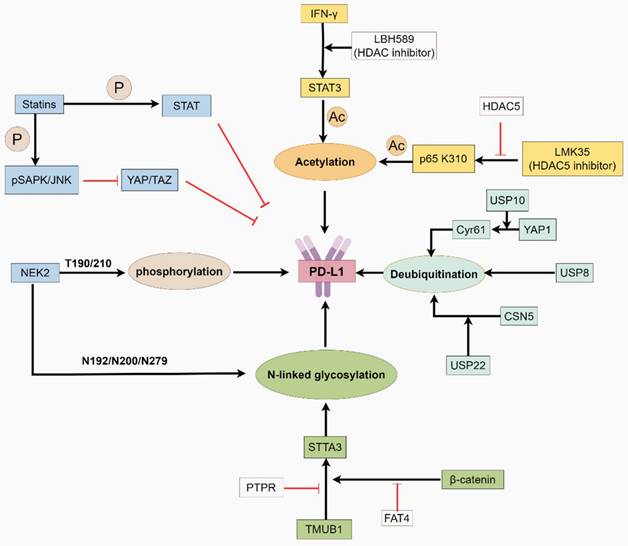
The half-life of PD-L1 is regulated by ubiquitination and deubiquitination [105]. DUBs alter substrate conformation by cleaving ubiquitin moieties, facilitating tumor immune evasion. DUBs regulate PD-L1 deubiquitination through distinct mechanisms. For instance, Ubiquitin-specific peptidase 10 (USP10) is a deubiquitinating enzyme that exhibits oncogenic effects in multiple tumors [106, 107]. YAP1, a key effector of the Hippo pathway, is involved in tumorigenesis and immunosuppression [107]. USP10 deubiquitinates and stabilizes YAP1/Cyr61, thereby increasing PD-L1 and galectin-9 in the TME and increasing the M2 macrophage proportion. This facilitates tumor invasion and immune evasion [108, 109]. In addition, Ubiquitin-specific peptidase 8 (USP8), another deubiquitinase, is associated with T-cell function [110]. USP8, a new PD-L1 deubiquitinase, interacts with PD-L1, thereby inhibiting its ubiquitination-dependent proteasomal degradation in pancreatic cancer. USP8 inhibitors combined with anti-PD-L1 therapy stimulate cytotoxic T cells and improve efficacy [111]. Ubiquitin-specific peptidase 22 (USP22) is overexpressed in various malignant tumors [112-114]. On the one hand, it directly regulates the stability of PD-L1 through ubiquitination. On the other hand, USP22 deubiquitinates COP9 signalosome subunit 5 (CSN5) and regulates PD-L1 protein levels through the USP22-CSN5-PD-L1 axis (Fig. 2) [114]. In the USP22 knockdown model, a decrease in M2 macrophage infiltration was also observed, indicating its multiple roles in immune regulation [115].
The E3 ubiquitin ligase determines the specificity of the ubiquitination reaction by identifying the substrate within the ubiquitin-protease system [116]. A growing body of research highlights the critical role of E3 ligases in modulating tumor immune responses [137]. E3 ubiquitin ligases RNF43 and ZNRF3 function as tumor suppressors in stem cell homeostasis by down-regulating Wnt receptors [117]. Single-cell sequencing revealed that RNF43-deficient tumor progression was accompanied by complex Immunological change, demonstrating low myeloid and high lymphocyte TME. The absence of RNF43 may result in the up-regulation of CTLA4 expression, potentially diminishing the efficacy of immunotherapy [118]. The linear ubiquitin chain assembly complex (LUBAC) can facilitate tumor progression in the TME [119]. NF31 inhibition, as a component of LUBAC, significantly enhances the sensitivity of tumor cells to NK- and T-cell-mediated killing. In vivo studies using tumor transplantation models have demonstrated that the impairment of RNF31 function results in diminished tumor growth and enhanced T-cell infiltration and efficacy [120].
The function of PD-L1 is regulated by post-translational modifications. This graphic was generated using Figdraw.
Sumoylation
Sumoylation is a dynamic and reversible PTM [121]. Initially discovered in yeast [122]. Five SUMO subtypes have been identified in humans [123]. The SUMOylation process involves an enzyme cascade comprising the SUMO E1 activating enzyme, E2 conjugating enzyme, E3 ligase, and deSUMOylating enzyme. SUMO-1 modifies substrates as a monomer, whereas SUMO-2/3 can form poly-SUMO chains [124]. SUMOylation is essential for regulating cellular functions, including protein activity, subcellular localization, and transcriptional regulation [123].
Emerging evidence highlights the significant role of SUMOylation in pancreatic cancer. SUMOylation affects PDAC adaptation and survival by regulating essential processes, including cell proliferation and migration [125]. Alexander Biederstädt et al. [126] found an aggressive pancreatic cancer subtype that co-actives MYC and SUMO pathways, which affect prognosis. Increased MYC activity increased PDAC sensitivity to SUMO inhibitors.
Previous studies have reported that mitotic SENP3 activation can lead to micronuclei formation in cancer cells and induce innate immunity through the cGAS-STING signaling pathway, thereby inducing host antitumor immunity [127, 128]. Analysis of pancreatic cancer samples from public databases revealed that SUMO1/2 expression is inversely associated with the infiltration of various tumor-infiltrating immune cells, including activated B cells, memory B cells, and effector memory CD8 T cells. This correlation encompasses most immune modulators, including chemokines, MHC molecules, immune promoters, and chemokine receptors [129]. Sumit Kumar et al. [129] have confirmed these findings. The SUMOylation inhibitor TAK-981 exhibits dual potential in PDAC treatment. It inhibits cell mitosis by targeting the SUMO pathway and simultaneously activates interferon signaling, enhancing CD8 T cells and NK cell infiltration.
Conclusion and Discussion
PTMs are critical events in signal transduction and are essential for regulating protein conformation, function, movement, and interactions. Beyond the classical PTMs discussed above, emerging modifications, including β-hydroxybutyrylation and lysine crotonylation, have been linked to tumor immune response and metabolism [130, 131].
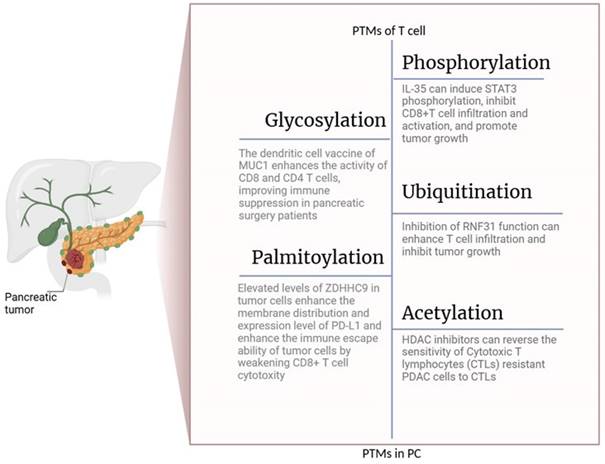
PTMs Regulate the Function of T Cells in pancreatic cancer. The figure is generated with BioRender (https://biorender.com).
Crotonylation is a modification that utilizes crotonyl-CoA as the donor [132]. In PDAC, crotonylation of metabolic enzymes significantly regulates tumor progression. For example, CBP/p300 facilitates IDH1 crotonylation at Lys224 and Lys236, which impacts metabolic levels. Besides, CBP/p300, combined with HDAC1 and HDAC3, facilitates MTHFD1 decrotonylation at K354 and K553, promoting pancreatic cancer development [133]. Investigating the mechanisms and functions of crotonylation in metabolic enzymes during PDAC progression may reveal new therapeutic targets.
Protein palmitoylation is a dynamic lipid modification facilitated by the ZDHHC protein family [134]. ZDHHC9 is significantly upregulated in pancreatic cancer than in normal tissues. Elevated levels of ZDHHC9 in tumor cells enhance the membrane distribution and expression level of PD-L1 and enhance the immune escape ability of tumor cells by weakening CD8+ T cell cytotoxicity (Fig. 3) [135].
Lysine 2-hydroxyisobutyrylation (Khib) is a new PTM found in histones that primarily regulates chromatin function [136]. KEGG analysis of Khib proteins reveals significant enrichment in glycolysis/gluconeogenesis pathways. Khib may significantly impact PDAC metabolism and facilitate tumor progression. The Khib protein inhibitor MG149 significantly inhibits PDAC migration and invasion, indicating that inhibitors targeting Khib proteins could be potential therapeutic targets for cancer treatment [135].
PTMs frequently interact to regulate protein functions instead of occurring independently. Emerging evidence highlights the extensive interaction among various PTMs in disease progression and treatment. O-GlcNAcylation, a PTM of SIRT7, can inhibit its interaction with REGγ and enhance histone deacetylation, consequently facilitating pancreatic cancer progression by preserving SIRT7 stability [137]. Mdm2, an E3 ubiquitin ligase, is the primary negative regulator of p53, facilitating its degradation through ubiquitination [138]. They possess overlapping acetylation sites, and acetylated p53 and Mdm2 repel each other, thus maintaining p53 stability and transcriptional activity [139, 140]. Understanding these PTM interactions can provide valuable insights into disease mechanisms and reveal new therapeutic targets.
PTMs are essential in the regulation of protein function, stability, interactions, and subcellular localization. Understanding the role of PTMs in the immune microenvironment and immunotherapy of pancreatic cancer can improve our comprehension of the disease and aid in developing new therapeutic strategies.
Abbreviations
PDAC: Pancreatic ductal adenocarcinoma
ACT: Adoptive cell therapy
CAR-T: Chimeric Antigen Receptor T-Cell
TCR-T: T-Cell Receptor-engineered T cell
TME: Tumor Microenvironment
MSI-H: Microsatellite Instability-High
PTM: Post-translational modification
SUMO: Small Ubiquitin-like Modifier
PD-1: Programmed Cell Death Protein 1
PD-L1: Programmed Death-Ligand 1
CTLA-4: Cytotoxic T-lymphocyte associated protein 4
NEK2: Never in Mitosis A-related kinase 2
GEM: Gemcitabine
STAT1: Signal Transducer and Activator of Transcription 1
Plk1: Polo-like Kinase 1
RB: retinoblastoma protein
TCR: T cell receptor TCR
TILs: tumor-infiltrating lymphocytes
TAMs: Tumor-associated macrophages
IL-35: Interleukin-35
HAS2: Hyaluronan synthase 2
DYRK1B: Dual-specificity tyrosine-regulated kinase 1B
IL-21: Interleukin-21
MUC4: Mucin 4
MUC1: Mucin 1
MGAT5: Glycosyltransferase 5
HDAC: Histone deacetylase
CTLs: Cytotoxic T lymphocytes
MDSC: Myeloid-derived suppressor cell
DUB: Deubiquitinating enzyme
USP10: Ubiquitin-specific peptidase 10
USP22: Ubiquitin-specific peptidase 22
USP8: Ubiquitin-specific peptidase 8
CSN5: COP9 signalosome subunit 5
LUBAC: linear ubiquitin chain assembly complex
Khib: Lysine 2-hydroxyisobutyrylation
Acknowledgements
We thank the members of this research group for their help and contributions.
Funding
This work was supported by The National Natural Science Foundation of China under Grant number 81772516, Provincial-level discipline leader of the NJPH under Grant number DTRB202213, Health projects of Yangzhou City Basic Research Plan (Joint Special Project) (grant NO. 2024-3-03), and Multi-discipline Scientific Research Foundation of Northern Jiangsu People's Hospital (grant NO. SBJC23004).
Author contributions
XG, KZ, JZ, YC, ZW, PW, PX, and JY contributed to the study conception and design and commented on previous versions of the manuscript. The first draft of the manuscript was written by XG. All authors contributed to the article and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605-17
2. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
3. Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16:11-26
4. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333-48
5. Ando Y, Mariano C, Shen K. Engineered in vitro tumor models for cell-based immunotherapy. Acta Biomater. 2021;132:345-59
6. Mathew D, Marmarelis ME, Foley C, Bauml JM, Ye D, Ghinnagow R. et al. Combined JAK inhibition and PD-1 immunotherapy for non-small cell lung cancer patients. Science. 2024;384:eadf1329
7. Ghaffari S, Khalili N, Rezaei N. CRISPR/Cas9 revitalizes adoptive T-cell therapy for cancer immunotherapy. J Exp Clin Cancer Res. 2021;40:269
8. Huang R, Li X, He Y, Zhu W, Gao L, Liu Y. et al. Recent advances in CAR-T cell engineering. J Hematol Oncol. 2020;13:86
9. Baulu E, Gardet C, Chuvin N, Depil S. TCR-engineered T cell therapy in solid tumors: State of the art and perspectives. Sci Adv. 2023;9:eadf3700
10. Zinn S, Vazquez-Lombardi R, Zimmermann C, Sapra P, Jermutus L, Christ D. Advances in antibody-based therapy in oncology. Nat Cancer. 2023;4:165-80
11. Johnson BA 3rd, Yarchoan M, Lee V, Laheru DA, Jaffee EM. Strategies for Increasing Pancreatic Tumor Immunogenicity. Clin Cancer Res. 2017;23:1656-69
12. Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell Localization, Activation, and Clonal Expansion in Human Pancreatic Ductal Adenocarcinoma. Cancer Immunol Res. 2017;5:978-91
13. Anderson KG, Stromnes IM, Greenberg PD. Obstacles Posed by the Tumor Microenvironment to T cell Activity: A Case for Synergistic Therapies. Cancer Cell. 2017;31:311-25
14. Balachandran VP, Beatty GL, Dougan SK. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology. 2019;156:2056-72
15. Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633-49
16. Ramazi S, Allahverdi A, Zahiri J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J Biosci. 2020;45:135
17. Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458-e1
18. Cui X, Wang J, Li K, Lv B, Hou B, Ding Z. Protein post-translational modifications in auxin signaling. J Genet Genomics. 2024;51:279-91
19. Hwang MS, Park J, Ham Y, Lee IH, Chun KH. Roles of Protein Post-Translational Modifications During Adipocyte Senescence. Int J Biol Sci. 2023;19:5245-56
20. Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A. et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590-6
21. Meng Y, Sandow JJ, Czabotar PE, Murphy JM. The regulation of necroptosis by post-translational modifications. Cell Death Differ. 2021;28:861-83
22. Patwardhan A, Cheng N, Trejo J. Post-Translational Modifications of G Protein-Coupled Receptors Control Cellular Signaling Dynamics in Space and Time. Pharmacol Rev. 2021;73:120-51
23. Yu Y, Liu J, Liu C, Liu R, Liu L, Yu Z. et al. Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation. Cells. 2022;11:3043
24. Kacen A, Javitt A, Kramer MP, Morgenstern D, Tsaban T, Shmueli MD. et al. Post-translational modifications reshape the antigenic landscape of the MHC I immunopeptidome in tumors. Nat Biotechnol. 2023;41:239-51
25. Kim EJ, Liu P, Zhang S, Donahue K, Wang Y, Schehr JL. et al. BAF155 methylation drives metastasis by hijacking super-enhancers and subverting anti-tumor immunity. Nucleic Acids Res. 2021;49:12211-33
26. Feng C, Zhang L, Chang X, Qin D, Zhang T. Regulation of post-translational modification of PD-L1 and advances in tumor immunotherapy. Front Immunol. 2023;14:1230135
27. Liu X, Zhang Y, Wang Y, Yang M, Hong F, Yang S. Protein Phosphorylation in Cancer: Role of Nitric Oxide Signaling Pathway. Biomolecules. 2021;11:1009
28. Xia S, Zhai Y, Wang X, Fan Q, Dong X, Chen M, Han T. Phosphorylation of polysaccharides: A review on the synthesis and bioactivities. Int J Biol Macromol. 2021;184:946-54
29. Yao S, Chen L. PD-1 as an immune modulatory receptor. Cancer J. 2014;20:262-4
30. Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019;76:359-70
31. Zhang X, Huang X, Xu J, Li E, Lao M, Tang T. et al. NEK2 inhibition triggers anti-pancreatic cancer immunity by targeting PD-L1. Nat Commun. 2021;12:4536
32. Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW. et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632
33. Doi T, Ishikawa T, Okayama T, Oka K, Mizushima K, Yasuda T. et al. The JAK/STAT pathway is involved in the upregulation of PD-L1 expression in pancreatic cancer cell lines. Oncol Rep. 2017;37:1545-54
34. Minz AP, Mohapatra D, Dutta M, Sethi M, Parida D, Mohapatra AP. et al. Statins abrogate gemcitabine-induced PD-L1 expression in pancreatic cancer-associated fibroblasts and cancer cells with improved therapeutic outcome. Cancer Immunol Immunother. 2023;72:4261-78
35. Uemura N, Hayashi H, Liu Z, Matsumura K, Ogata Y, Yasuda N. et al. Statins exert anti-growth effects by suppressing YAP/TAZ expressions via JNK signal activation and eliminate the immune suppression by downregulating PD-L1 expression in pancreatic cancer. Am J Cancer Res. 2023;13:2041-54
36. Wang H, Zhang H, Wang Y, Brown ZJ, Xia Y, Huang Z. et al. Regulatory T-cell and neutrophil extracellular trap interaction contributes to carcinogenesis in non-alcoholic steatohepatitis. J Hepatol. 2021;75:1271-83
37. Kaltenmeier C, Yazdani HO, Morder K, Geller DA, Simmons RL, Tohme S. Neutrophil Extracellular Traps Promote T Cell Exhaustion in the Tumor Microenvironment. Front Immunol. 2021;12:785222
38. Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L. et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2016;29:832-45
39. Bailly C. Toward a repositioning of the antibacterial drug nifuroxazide for cancer treatment. Drug Discov Today. 2019;24:1930-6
40. Xie Y, Zhou T, Li X, Zhao K, Bai W, Hou X. et al. Targeting ESE3/EHF With Nifurtimox Inhibits CXCR2(+) Neutrophil Infiltration and Overcomes Pancreatic Cancer Resistance to Chemotherapy and Immunotherapy. Gastroenterology. 2024;167:281-97
41. Zhang Z, Cheng L, Li J, Qiao Q, Karki A, Allison DB. et al. Targeting Plk1 Sensitizes Pancreatic Cancer to Immune Checkpoint Therapy. Cancer Res. 2022;82:3532-48
42. Kruusmägi M, Zelenin S, Brismar H, Scott L. Intracellular dynamics of calcyon, a neuron-specific vesicular protein. Neuroreport. 2007;18:1547-51
43. Xia X, Li R, Zhou P, Xing Z, Lu C, Long Z. et al. Decreased NSG3 enhances PD-L1 expression by Erk1/2 pathway to promote pancreatic cancer progress. Am J Cancer Res. 2021;11:916-29
44. Shi D, Shi Y, Kaseb AO, Qi X, Zhang Y, Chi J. et al. Chimeric Antigen Receptor-Glypican-3 T-Cell Therapy for Advanced Hepatocellular Carcinoma: Results of Phase I Trials. Clin Cancer Res. 2020;26:3979-89
45. Zhang B, Wang CM, Wu HX, Wang F, Chai YY, Hu Y. et al. MFSD2A potentiates gastric cancer response to anti-PD-1 immunotherapy by reprogramming the tumor microenvironment to activate T cell response. Cancer Commun (Lond). 2023;43:1097-116
46. Somani VK, Zhang D, Dodhiawala PB, Lander VE, Liu X, Kang LI. et al. IRAK4 Signaling Drives Resistance to Checkpoint Immunotherapy in Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2022;162:2047-62
47. Liu A, Liu Y, Li PK, Li C, Lin J. LLL12 inhibits endogenous and exogenous interleukin-6-induced STAT3 phosphorylation in human pancreatic cancer cells. Anticancer Res. 2011;31:2029-35
48. Alam MS, Gaida MM, Ogawa Y, Kolios AG, Lasitschka F, Ashwell JD. Counter-regulation of T cell effector function by differentially activated p38. J Exp Med. 2014;211:1257-70
49. Alam MS, Gaida MM, Bergmann F, Lasitschka F, Giese T, Giese NA. et al. Selective inhibition of the p38 alternative activation pathway in infiltrating T cells inhibits pancreatic cancer progression. Nat Med. 2015;21:1337-43
50. Mirlekar B, Michaud D, Lee SJ, Kren NP, Harris C, Greene K. et al. B cell-Derived IL35 Drives STAT3-Dependent CD8(+) T-cell Exclusion in Pancreatic Cancer. Cancer Immunol Res. 2020;8:292-308
51. Mirlekar B, Michaud D, Searcy R, Greene K, Pylayeva-Gupta Y. IL35 Hinders Endogenous Antitumor T-cell Immunity and Responsiveness to Immunotherapy in Pancreatic Cancer. Cancer Immunol Res. 2018;6:1014-24
52. Zhao T, Ren H, Jia L, Chen J, Xin W, Yan F. et al. Inhibition of HIF-1α by PX-478 enhances the anti-tumor effect of gemcitabine by inducing immunogenic cell death in pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:2250-62
53. Zhang Y, Velez-Delgado A, Mathew E, Li D, Mendez FM, Flannagan K. et al. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut. 2017;66:124-36
54. Lee BY, Hogg EKJ, Below CR, Kononov A, Blanco-Gomez A, Heider F. et al. Heterocellular OSM-OSMR signalling reprograms fibroblasts to promote pancreatic cancer growth and metastasis. Nat Commun. 2021;12:7336
55. He Z, Wang J, Zhu C, Xu J, Chen P, Jiang X. et al. Exosome-derived FGD5-AS1 promotes tumor-associated macrophage M2 polarization-mediated pancreatic cancer cell proliferation and metastasis. Cancer Lett. 2022;548:215751
56. Georgoudaki AM, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J. et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016;15:2000-11
57. Friedman E. The Kinase Mirk/dyrk1B: A Possible Therapeutic Target in Pancreatic Cancer. Cancers (Basel). 2010;2:1492-512
58. Brichkina A, Ems M, Suezov R, Singh R, Lutz V, Picard FSR. et al. DYRK1B blockade promotes tumoricidal macrophage activity in pancreatic cancer. Gut. 2024;73:1684-1701
59. Blom K, Rubin J, Berglund M, Jarvius M, Lenhammar L, Parrow V. et al. Mebendazole-induced M1 polarisation of THP-1 macrophages may involve DYRK1B inhibition. BMC Res Notes. 2019;12:234
60. Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234-48
61. Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH. et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71:5020-9
62. Caligiuri MA. Human natural killer cells. Blood. 2008;112:461-9
63. Liu S, Galat V, Galat Y, Lee YKA, Wainwright D, Wu J. NK cell-based cancer immunotherapy: from basic biology to clinical development. J Hematol Oncol. 2021;14:7
64. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity. 2017;47:820-33
65. Walle T, Kraske JA, Liao B, Lenoir B, Timke C, von Bohlen Und Halbach E. et al. Radiotherapy orchestrates natural killer cell dependent antitumor immune responses through CXCL8. Sci Adv. 2022;8:eabh4050
66. Xie X, Zhou Y, Wang X, Guo J, Li J, Fan H. et al. Enhanced antitumor activity of gemcitabine by polysaccharide-induced NK cell activation and immune cytotoxicity reduction in vitro/vivo. Carbohydr Polym. 2017;173:360-71
67. Xie X, Ma L, Zhou Y, Shen W, Xu D, Dou J. et al. Polysaccharide enhanced NK cell cytotoxicity against pancreatic cancer via TLR4/MAPKs/NF-κB pathway in vitro/vivo. Carbohydr Polym. 2019;225:115223
68. Rezvani K, Rouce RH. The Application of Natural Killer Cell Immunotherapy for the Treatment of Cancer. Front Immunol. 2015;6:578
69. McMichael EL, Jaime-Ramirez AC, Guenterberg KD, Luedke E, Atwal LS, Campbell AR. et al. IL-21 Enhances Natural Killer Cell Response to Cetuximab-Coated Pancreatic Tumor Cells. Clin Cancer Res. 2017;23:489-502
70. Thomas D, Rathinavel AK, Radhakrishnan P. Altered glycosylation in cancer: A promising target for biomarkers and therapeutics. Biochim Biophys Acta Rev Cancer. 2021;1875:188464
71. Tang H, Partyka K, Hsueh P, Sinha JY, Kletter D, Zeh H. et al. Glycans related to the CA19-9 antigen are elevated in distinct subsets of pancreatic cancers and improve diagnostic accuracy over CA19-9. Cell Mol Gastroenterol Hepatol. 2016;2:201-21.e15
72. Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, Haab BB. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697-707
73. Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6:1126-38
74. Mereiter S, Balmaña M, Gomes J, Magalhães A, Reis CA. Glycomic Approaches for the Discovery of Targets in Gastrointestinal Cancer. Front Oncol. 2016;6:55
75. Rodriguez E, Boelaars K, Brown K, Eveline Li RJ, Kruijssen L, Bruijns SCM. et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat Commun. 2021;12:1270
76. Costa AF, Campos D, Reis CA, Gomes C. Targeting Glycosylation: A New Road for Cancer Drug Discovery. Trends Cancer. 2020;6:757-66
77. Lübbers J, Rodríguez E, van Kooyk Y. Modulation of Immune Tolerance via Siglec-Sialic Acid Interactions. Front Immunol. 2018;9:2807
78. Perdicchio M, Cornelissen LA, Streng-Ouwehand I, Engels S, Verstege MI, Boon L. et al. Tumor sialylation impedes T cell mediated anti-tumor responses while promoting tumor associated-regulatory T cells. Oncotarget. 2016;7:8771-82
79. Büll C, Boltje TJ, Balneger N, Weischer SM, Wassink M, van Gemst JJ. et al. Sialic Acid Blockade Suppresses Tumor Growth by Enhancing T-cell-Mediated Tumor Immunity. Cancer Res. 2018;78:3574-88
80. Wei J, Gao W, Wu J, Meng K, Zhang J, Chen J, Miao Y. Dendritic cells expressing a combined PADRE/MUC4-derived polyepitope DNA vaccine induce multiple cytotoxic T-cell responses. Cancer Biother Radiopharm. 2008;23:121-8
81. Cai H, Palitzsch B, Hartmann S, Stergiou N, Kunz H, Schmitt E, Westerlind U. Antibody induction directed against the tumor-associated MUC4 glycoprotein. Chembiochem. 2015;16:959-67
82. Lepisto AJ, Moser AJ, Zeh H, Lee K, Bartlett D, McKolanis JR. et al. A phase I/II study of a MUC1 peptide pulsed autologous dendritic cell vaccine as adjuvant therapy in patients with resected pancreatic and biliary tumors. Cancer Ther. 2008;6:955-64
83. Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat Med. 2019;25:1341-55
84. Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L. et al. Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discov. 2020;10:702-23
85. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540-55
86. Hollander EE, Flock RE, McDevitt JC, Vostrejs WP, Campbell SL, Orlen MI. et al. N-glycosylation by Mgat5 imposes a targetable constraint on immune-mediated tumor clearance. JCI Insight. 2024;9:e178804
87. Greco B, Malacarne V, De Girardi F, Scotti GM, Manfredi F, Angelino E. et al. Disrupting N-glycan expression on tumor cells boosts chimeric antigen receptor T cell efficacy against solid malignancies. Sci Transl Med. 2022;14:eabg3072
88. Bartish M, Del Rincón SV, Rudd CE, Saragovi HU. Aiming for the Sweet Spot: Glyco-Immune Checkpoints and γδ T Cells in Targeted Immunotherapy. Front Immunol. 2020;11:564499
89. Posey AD Jr, Schwab RD, Boesteanu AC, Steentoft C, Mandel U, Engels B. et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity. 2016;44:1444-54
90. Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125-31
91. Allfrey VG, Faulkner R, Mirsky AE. ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proc Natl Acad Sci U S A. 1964;51:786-94
92. Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595-606
93. Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T. et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633-6
94. Blaszczak W, Liu G, Zhu H, Barczak W, Shrestha A, Albayrak G. et al. Immune modulation underpins the anti-cancer activity of HDAC inhibitors. Mol Oncol. 2021;15:3280-98
95. West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30-9
96. Wang X, Zhao J. Targeted Cancer Therapy Based on Acetylation and Deacetylation of Key Proteins Involved in Double-Strand Break Repair. Cancer Manag Res. 2022;14:259-71
97. Zhou Y, Jin X, Yu H, Qin G, Pan P, Zhao J. et al. HDAC5 modulates PD-L1 expression and cancer immunity via p65 deacetylation in pancreatic cancer. Theranostics. 2022;12:2080-94
98. Woods DM, Sodré AL, Villagra A, Sarnaik A, Sotomayor EM, Weber J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol Res. 2015;3:1375-85
99. Hu G, He N, Cai C, Cai F, Fan P, Zheng Z, Jin X. HDAC3 modulates cancer immunity via increasing PD-L1 expression in pancreatic cancer. Pancreatology. 2019;19:383-9
100. Looi CK, Gan LL, Sim W, Hii LW, Chung FF, Leong CO. et al. Histone Deacetylase Inhibitors Restore Cancer Cell Sensitivity towards T Lymphocytes Mediated Cytotoxicity in Pancreatic Cancer. Cancers (Basel). 2022;14:3709
101. Christmas BJ, Rafie CI, Hopkins AC, Scott BA, Ma HS, Cruz KA. et al. Entinostat Converts Immune-Resistant Breast and Pancreatic Cancers into Checkpoint-Responsive Tumors by Reprogramming Tumor-Infiltrating MDSCs. Cancer Immunol Res. 2018;6:1561-77
102. Wu X, Xu M, Geng M, Chen S, Little PJ, Xu S, Weng J. Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies. Signal Transduct Target Ther. 2023;8:220
103. Hu X, Wang J, Chu M, Liu Y, Wang ZW, Zhu X. Emerging Role of Ubiquitination in the Regulation of PD-1/PD-L1 in Cancer Immunotherapy. Mol Ther. 2021;29:908-19
104. Garcia-Sanchez JA, Ewbank JJ, Visvikis O. Ubiquitin-related processes and innate immunity in C. elegans. Cell Mol Life Sci. 2021;78:4305-33
105. Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19:59-70
106. Hu C, Zhang M, Moses N, Hu CL, Polin L, Chen W. et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020;11:328
107. Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S. et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197-210
108. Xia P, Liu P, Fu Q, Liu C, Luo Q, Zhang X. et al. Long noncoding RNA EPIC1 interacts with YAP1 to regulate the cell cycle and promote the growth of pancreatic cancer cells. Biochem Biophys Res Commun. 2020;522:978-85
109. Liu X, Chen B, Chen J, Su Z, Sun S. Deubiquitinase ubiquitin-specific peptidase 10 maintains cysteine rich angiogenic inducer 61 expression via Yes1 associated transcriptional regulator to augment immune escape and metastasis of pancreatic adenocarcinoma. Cancer Sci. 2022;113:1868-79
110. Dufner A, Kisser A, Niendorf S, Basters A, Reissig S, Schönle A. et al. The ubiquitin-specific protease USP8 is critical for the development and homeostasis of T cells. Nat Immunol. 2015;16:950-60
111. Yang H, Zhang X, Lao M, Sun K, He L, Xu J. et al. Targeting ubiquitin-specific protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy of pancreatic cancer. Cell Death Differ. 2023;30:560-75
112. McCann JJ, Vasilevskaya IA, Poudel Neupane N, Shafi AA, McNair C, Dylgjeri E. et al. USP22 Functions as an Oncogenic Driver in Prostate Cancer by Regulating Cell Proliferation and DNA Repair. Cancer Res. 2020;80:430-43
113. Bai Z, Du Y, Cong L, Cheng Y. The USP22 promotes the growth of cancer cells through the DYRK1A in pancreatic ductal adenocarcinoma. Gene. 2020;758:144960
114. Prokakis E, Bamahmoud H, Jansari S, Fritsche L, Dietz A, Boshnakovska A. et al. USP22 supports the aggressive behavior of basal-like breast cancer by stimulating cellular respiration. Cell Commun Signal. 2024;22:120
115. Huang J, Yin Q, Wang Y, Zhou X, Guo Y, Tang Y. et al. EZH2 Inhibition Enhances PD-L1 Protein Stability Through USP22-Mediated Deubiquitination in Colorectal Cancer. Adv Sci (Weinh). 2024;11:e2308045
116. Zhou X, Sun SC. Targeting ubiquitin signaling for cancer immunotherapy. Signal Transduct Target Ther. 2021;6:16
117. Farnhammer F, Colozza G, Kim J. RNF43 and ZNRF3 in Wnt Signaling - A Master Regulator at the Membrane. Int J Stem Cells. 2023;16:376-84
118. Hosein AN, Dangol G, Okumura T, Roszik J, Rajapakshe K, Siemann M. et al. Loss of Rnf43 Accelerates Kras-Mediated Neoplasia and Remodels the Tumor Immune Microenvironment in Pancreatic Adenocarcinoma. Gastroenterology. 2022;162:1303-18.e18
119. Zhang J, Tu H, Zheng Z, Zhao X, Lin X. RNF31 promotes tumorigenesis via inhibiting RIPK1 kinase-dependent apoptosis. Oncogene. 2023;42:1585-96
120. Zhang Z, Kong X, Ligtenberg MA, van Hal-van Veen SE, Visser NL, de Bruijn B. et al. RNF31 inhibition sensitizes tumors to bystander killing by innate and adaptive immune cells. Cell Rep Med. 2022;3:100655
121. Ryu HY, Ahn SH, Hochstrasser M. SUMO and cellular adaptive mechanisms. Exp Mol Med. 2020;52:931-9
122. Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793-807
123. Celen AB, Sahin U. Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. Febs j. 2020;287:3110-40
124. Chang HM, Yeh ETH. SUMO: From Bench to Bedside. Physiol Rev. 2020;100:1599-619
125. Rauth S, Karmakar S, Shah A, Seshacharyulu P, Nimmakayala RK, Ganguly K. et al. SUMO Modification of PAF1/PD2 Enables PML Interaction and Promotes Radiation Resistance in Pancreatic Ductal Adenocarcinoma. Mol Cell Biol. 2021;41:e0013521
126. Biederstädt A, Hassan Z, Schneeweis C, Schick M, Schneider L, Muckenhuber A. et al. SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut. 2020;69:1472-82
127. Wei B, Huang C, Liu B, Wang Y, Xia N, Fan Q. et al. Mitotic Phosphorylation of SENP3 Regulates DeSUMOylation of Chromosome-Associated Proteins and Chromosome Stability. Cancer Res. 2018;78:2171-8
128. Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A. et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843-52
129. Kumar S, Schoonderwoerd MJA, Kroonen JS, de Graaf IJ, Sluijter M, Ruano D. et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. 2022;71:2266-83
130. Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457-70
131. Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137-42
132. Wan J, Liu H, Chu J, Zhang H. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019;23:7163-9
133. Zheng Y, Zhu L, Qin ZY, Guo Y, Wang S, Xue M. et al. Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression. Cell Rep. 2023;42:112666
134. Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74-84
135. Lin Z, Huang K, Guo H, Jia M, Sun Q, Chen X. et al. Targeting ZDHHC9 potentiates anti-programmed death-ligand 1 immunotherapy of pancreatic cancer by modifying the tumor microenvironment. Biomed Pharmacother. 2023;161:114567
136. Huang S, Tang D, Dai Y. Metabolic Functions of Lysine 2-Hydroxyisobutyrylation. Cureus. 2020;12:e9651
137. He X, Li Y, Chen Q, Zheng L, Lou J, Lin C. et al. O-GlcNAcylation and stablization of SIRT7 promote pancreatic cancer progression by blocking the SIRT7-REGγ interaction. Cell Death Differ. 2022;29:1970-81
138. Feeley KP, Adams CM, Mitra R, Eischen CM. Mdm2 Is Required for Survival and Growth of p53-Deficient Cancer Cells. Cancer Res. 2017;77:3823-33
139. Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. Embo j. 2001;20:1331-40
140. Stehmeier P, Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst). 2009;8:491-8
141. Zhou C, Zhu X, Liu N, Dong X, Zhang X, Huang H. et al. B-lymphoid tyrosine kinase-mediated FAM83A phosphorylation elevates pancreatic tumorigenesis through interacting with β-catenin. Signal Transduct Target Ther. 2023;8:66
142. Jeong MH, Urquhart G, Lewis C, Chi Z, Jewell JL. Inhibition of phosphodiesterase 4D suppresses mTORC1 signaling and pancreatic cancer growth. JCI Insight. 2023;8:e158098
143. Yang L, Fu Q, Miao L, Ding Q, Li X, Wang J. et al. Quantitative acetylome and phosphorylome analysis reveals Girdin affects pancreatic cancer progression through regulating Cortactin. Aging (Albany NY). 2020;12:7679-93
144. Xiao Y, Chen Y, Chen J, Dong J. ASPP2 Is Phosphorylated by CDK1 during Mitosis and Required for Pancreatic Cancer Cell Proliferation. Cancers (Basel). 2023;15:5424
145. Huang S, Huang C, Chen W, Liu Y, Yin X, Lai J. et al. WAVE3 promotes proliferation, migration and invasion via the AKT pathway in pancreatic cancer. Int J Oncol. 2018;53:672-84
146. Garcia MN, Grasso D, Lopez-Millan MB, Hamidi T, Loncle C, Tomasini R. et al. IER3 supports KRASG12D-dependent pancreatic cancer development by sustaining ERK1/2 phosphorylation. J Clin Invest. 2014;124:4709-22
147. Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013;10:607-20
148. Guo H, Xiao Y, Yuan Z, Yang X, Chen J, Chen C. et al. Inhibition of STAT3(Y705) phosphorylation by Stattic suppresses proliferation and induces mitochondrial-dependent apoptosis in pancreatic cancer cells. Cell Death Discov. 2022;8:116
149. Wu H, Hasan R, Zhang H, Gray J, Williams D, Miller M. et al. Phosphorylation Regulates CAP1 (Cyclase-Associated Protein 1) Functions in the Motility and Invasion of Pancreatic Cancer Cells. Sci Rep. 2019;9:4925
150. Xiao Y, Chen Y, Peng A, Dong J. The phosphatase CTDSPL2 is phosphorylated in mitosis and a target for restraining tumor growth and motility in pancreatic cancer. Cancer Lett. 2022;526:53-65
151. Song C, Wang G, Liu M, Han S, Dong M, Peng M. et al. Deciphering the SOX4/MAPK1 regulatory axis: a phosphoproteomic insight into IQGAP1 phosphorylation and pancreatic Cancer progression. J Transl Med. 2024;22:602
152. Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H. et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9:1307-11
153. Bose M, Sanders A, De C, Zhou R, Lala P, Shwartz S. et al. Targeting tumor-associated MUC1 overcomes anoikis-resistance in pancreatic cancer. Transl Res. 2023;253:41-56
154. Luo G, Jin K, Deng S, Cheng H, Fan Z, Gong Y. et al. Roles of CA19-9 in pancreatic cancer: Biomarker, predictor and promoter. Biochim Biophys Acta Rev Cancer. 2021;1875:188409
155. Zhu Q, Zhou H, Wu L, Lai Z, Geng D, Yang W. et al. O-GlcNAcylation promotes pancreatic tumor growth by regulating malate dehydrogenase 1. Nat Chem Biol. 2022;18:1087-95
156. Leon F, Seshacharyulu P, Nimmakayala RK, Chugh S, Karmakar S, Nallasamy P. et al. Reduction in O-glycome induces differentially glycosylated CD44 to promote stemness and metastasis in pancreatic cancer. Oncogene. 2022;41:57-71
157. Holdbrooks AT, Britain CM, Bellis SL. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 2018;293:1610-22
158. Hu T, Shukla SK, Vernucci E, He C, Wang D, King RJ. et al. Metabolic Rewiring by Loss of Sirt5 Promotes Kras-Induced Pancreatic Cancer Progression. Gastroenterology. 2021;161:1584-600
159. Wang Y, Liu Y, Wang C, Kang R, Tang D, Liu J. EP300 promotes ferroptosis via HSPA5 acetylation in pancreatic cancer. Sci Rep. 2023;13:15004
160. Lei MZ, Li XX, Zhang Y, Li JT, Zhang F, Wang YP. et al. Acetylation promotes BCAT2 degradation to suppress BCAA catabolism and pancreatic cancer growth. Signal Transduct Target Ther. 2020;5:70
161. Huang X, Pan L, Zuo Z, Li M, Zeng L, Li R. et al. LINC00842 inactivates transcription co-regulator PGC-1α to promote pancreatic cancer malignancy through metabolic remodelling. Nat Commun. 2021;12:3830
162. He Y, Han P, Chen C, Xie S, Zhang H, Song Y. et al. circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation. Cancer Gene Ther. 2023;30:559-66
163. Nelson JK, Thin MZ, Evan T, Howell S, Wu M, Almeida B. et al. USP25 promotes pathological HIF-1-driven metabolic reprogramming and is a potential therapeutic target in pancreatic cancer. Nat Commun. 2022;13:2070
164. Wang Y, Sun Q, Mu N, Sun X, Wang Y, Fan S. et al. The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells. Cell Commun Signal. 2020;18:112
165. Lin S. DTX3L mediated ubiquitination of cGAS suppresses antitumor immunity in pancreatic cancer. Biochem Biophys Res Commun. 2023;681:106-10
166. Kang HW, Kim JH, Lee DE, Lee YS, Kim MJ, Kim HS. et al. Combination therapy of niclosamide with gemcitabine inhibited cell proliferation and apoptosis via Wnt/β-catenin/c-Myc signaling pathway by inducing β-catenin ubiquitination in pancreatic cancer. Cancer Biol Ther. 2023;24:2272334
167. Su D, Wang R, Chen G, Ding C, Liu Y, Tao J. et al. FBXO32 Stimulates Protein Synthesis to Drive Pancreatic Cancer Progression and Metastasis. Cancer Res. 2024;84:2607-25
Author contact
![]() Corresponding authors: Peng Xu, pengxu1986com; Jie Yao, dryaojieedu.cn.
Corresponding authors: Peng Xu, pengxu1986com; Jie Yao, dryaojieedu.cn.

 Global reach, higher impact
Global reach, higher impact