10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(1):43-52. doi:10.7150/ijbs.6818 This issue Cite
Research Paper
Important Functional Roles of Basigin in Thymocyte Development and T cell Activation
1. Cell Engineering Research Centre & Department of Cell Biology, State Key Laboratory of Cancer Biology, Fourth Military Medical University, 169 West Changle Street, Xi'an 710032, China.
2. Genetic Laboratory of Development and Disease, Institute of Biotechnology, 20 Dongdajie, Fengtai District, Beijing 100071, China.
* These authors contributed equally to this work.
Received 2013-6-2; Accepted 2013-11-22; Published 2013-12-13
Abstract
Basigin is a highly glycosylated transmembrane protein that is expressed in a broad range of tissues and is involved in a number of physiological and pathological processes. However, the in vivo role of basigin remains unknown. To better understand the physiological and pathological functions of basigin in vivo, we generated a conditional null allele by introducing two loxP sites flanking exons 2 and 7 of the basigin gene (Bsg). Bsgfl/fl mice were born at the expected Mendelian ratio and showed a similar growth rate compared with wildtype mice. After crossing these mice with Lck-Cre transgenic mice, basigin expression was specifically inactivated in T cells in the resulting Lck-Cre; Bsgfl/fl mice. Although the birth and growth rate of Lck-Cre; Bsgfl/fl mice were similar to control mice, thymus development was partially arrested in Lck-Cre; Bsgfl/fl mice, specifically at the CD4+CD8+ double-positive (DP) and CD4 single-positive (CD4+CD8-, CD4SP) stages. In addition, CD4+ T cell activation was enhanced upon Concanavalin A (Con A) or anti-CD3/anti-CD28 stimulation but not upon PMA/Ionomycin stimulation in the absence of basigin. Overall, this study provided the first in vivo evidence for the function of basigin in thymus development. Moreover, the successful generation of the conditional null basigin allele provides a useful tool for the study of distinct physiological or pathological functions of basigin in different tissues at different development stages.
Keywords: Basigin, Cre-Loxp, gene targeting, thymocyte development, T cell activation
Introduction
Basigin, which is also known as CD147 or EMMPRIN (extracellular matrix metalloproteinase inducer), encodes a transmembrane glycoprotein with two immunoglobulin-like domains [1, 2]. Basigin is expressed in various tissues and is involved in a number of physiological processes, including spermatogenesis [3, 4], fertilization [5], embryo implantation [3, 5], and neural functions, such as vision, behavior, memory, and olfaction [6-8]. Within the thymus, the expression level of basigin correlates with various differentiation stages of thymocytes, with the highest expression level found in early immature thymocytes. Treatment of immature fetal thymocytes using an anti-basigin monoclonal antibody (mAb) blocked thymocyte maturation ex vivo [9]. In adults, basigin was identified as a lymphocyte activation-associated antigen [10]. Moreover, its expression was also significantly upregulated in activated T lymphocytes. MAbs specific for basigin inhibited anti-CD3-induced T lymphocyte activation, presumably via the inhibition of the reorganization and clustering of GPI-anchored co-receptors within lipid rafts, which, in turn, altered downstream signaling events [11]. Our previous study showed that basigin was functionally linked to the formation of the immunological synapse [12]. However, the precise function and mechanism of basigin in regulating thymus development and T cell functions in vivo remains to be clarified. Basigin is also expressed in a large number of pathological conditions, including inflammation [13, 14], tissue repair/remodeling [15], ischemic disease [16, 17] and Alzheimer's disease [18]. It has been well established that basigin is overexpressed on the surface of tumor cells, and plays significant roles in tumor growth, angiogenesis, invasion, and metastasis by inducing the secretion of various matrix metalloproteinases [19], stimulating the production of vascular endothelial growth factor [20], activating cell signaling pathways [21, 22], preventing anoikis [23], and regulating metabolism [24], among others. Furthermore, overexpression of basigin in tumor tissues is correlated with the poor prognosis of patients with several types of solid tumors [25-28]. These results indicated that basigin is a widely expressed multifunctional protein and is implicated in a variety of physiological and pathological conditions.
Although basigin has been recognized to be a multifunctional protein, the molecular mechanisms underlying its diverse functions have yet to be elucidated. To examine the functions of basigin in vivo, Igakura et al. generated a basigin (Bsg) null mutant mouse (Bsg-/-) using a conventional gene targeting strategy [3]. Using this mouse model, they revealed that basigin played a critical role in embryo implantation, reproduction, physiological neural functions, and the mitogenic response of lymphocytes upon a mixed lymphocytic response [4, 5, 29]. However, because most of the Bsg-/- embryos died around the time of implantation and only a small number of Bsg-/- embryos survived until birth and into adulthood, it was very difficult to obtain a sufficient number of Bsg-/- mice for a more detailed phenotypic analysis. Moreover, the Bsg-/- mice that did survive into adulthood were smaller and weaker compared to wildtype controls. In addition, both male and female Bsg-/- mice were infertile, which prevented the establishment of several disease models for the study of the functions of basigin in a pathological context. Furthermore, knocking out the expression of basigin ubiquitously in all tissues prevented the ability to study its roles in specific tissues or at specific developmental stages, as well as the corresponding cellular and molecular mechanisms.
To overcome these disadvantages, we generated a conditional null allele for basigin using the Cre-loxP system. We also ablated the basigin gene in thymocytes and T lymphocytes using Lck-Cre transgenic mice. We found that Lck-Cre; Bsgfl/fl mice exhibited a significant reduction in the number of DP (double positive, CD4+CD8+) and CD4SP (single positive, CD4+CD8-) thymocytes, and a high proliferation index of CD4+ T cells upon Concanavalin A (Con A) or anti-CD3/CD28 stimulation. To the best of our knowledge, this is the first study to provide insight into the in vivo role of basigin in thymocytic development. Moreover, basigin conditional null mice will be a valuable genetic tool used to uncover the in vivo role of basigin in specific tissues and at specific developmental stages.
Materials and Methods
Construction of the Targeting Vector
The basigin conditional gene targeting vector was generated based on a PCR-based cloning strategy as previously described [30]. Basigin genomic DNA isolated from R1 ES cells (on 129/SV background) was used. To generate the targeting vector, the basigin fragment containing exons 2-7, a 1.5-kb fragment of the 5' arm and a 3.7-kb fragment of the 3' arm were amplified using PCR. The following PCR primers were synthesized by Invitrogen and used in this study: Basigin fragment with exons 2-7 primers: 5'-GTCGACCTTGTAGTAACGGGTACTAACCCTT-3' and 5'-ATCGATGACACACACATTGAGTCCCAGAGCA-3'; 5' arm primers: 5'-AAGGAAAAAAGCGGCCGCAGGCTGAATTTGATATTAGGGTCTC-3' and 5'-CCGCTCGAGCTCCATTTCTTTTCTGCTTGCGGGG-3'. 3' arm primers: 5'-CCCATCGATAGATCTATAACTTCGTATAATGTATGCTATACGAAGTTATAGGTGGATGGCTGCTGTTGAAATAA-3' and 5'-CGGGGTACCCAGTTAATCAATGGTTGATCAATCG-3'. The forward primer of the 3' arm was designed to introduce a Bgl II site and a loxP site into the PCR product. High-fidelity Taq polymerase (TaKaRa) was used for PCR amplification and all of the PCR products were confirmed by sequencing. These fragments were sequentially subcloned into the pfrtI vector (developed in our lab). A schematic of the conditional basigin targeting vector is shown in Fig. 1A.
Generation of Mice Carrying a conditional null allele for basigin
After linearization by Not I (TaKaRa), the targeting vector was transfected into R1 ES cells. Colonies resistant to G418 and ganciclovir were screened by Southern blot and PCR. Correctly targeted ES cells were then microinjected into C57BL/6 blastocysts to generate chimeric mice, which were further bred to C57BL/6 mice to generate F1 offspring. The conditional basigin allele (Bsgfn) mice were bred with Flp transgenic mice to delete the FRT flanked Neo cassette to produce Bsgfl mice, which were then bred with Lck-Cre transgenic mice to generate Lck-Cre; Bsgfl/fl mice. All of the mice used in this study were housed in a specific pathogen-free facility. Moreover, all of the animal work was approved by the Fourth Military Medical University's institutional animal care and use committee and handled in strict accordance with good animal practice as defined by the relevant national animal welfare bodies.
Genotypic analyses
Mouse genotypes were analyzed using Southern blot and PCR. Genomic DNA from ES cells and tissues were prepared according to standard protocols. A Southern blot was performed using a digoxigenin-based labeling and detection kit (Roche) according to the manufacturer's instructions. The 5' arm homologous recombination was identified by PCR using a Neo-specific primer pair, P1: 5'-ACAGACCCATAGCCTACAGC-3' and P2: 5'-GCTACTTCCATTTGTCACGT-3', expected to amplify a fragment of 2.1 kb in Bsgfn alleles. The loxP site flanked exon 7 was identified using a primer pair, P3: 5'-CTCTGGGACTCAATGTGTGT-3' and P4: 5'-AGGTGGGTTTTCTGTAAGGT-3', which was expected to amplify a 371-bp fragment in the wildtype allele and a 405-bp fragment in the targeted allele. The Flp-mediated deletion of the Neo cassette was identified using the primer pair, P5: 5'-CTGGAACTCCTAGCAATC-3' and P6: 5'-TGGGAAAGGGTTAGTAC-3', which was expected to amplify a 445-bp fragment in the Neo-deleted allele (Bsgfl) and a 377-bp fragment in the wildtype allele. The Cre-mediated deletion of the floxed exons 2 to 7 was identified using the primer pair, P7: 5'-CTGGAACTCCTAGCAATC-3' and P4, which was expected to amplify a 614-bp fragment (referred to as the deleted band). All of the PCR reactions were performed for 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 0.5 or 2 minutes.
Cell isolation
Spleens and thymuses were harvested from five-week-old mice and mechanically disrupted through a 70-µm strainer. After treatment with red blood cell lysis buffer (0.14 M NH4Cl in HEPES, pH 7.0), the cells were washed with a sterile phosphate buffer solution (PBS) and resuspended as a single cell suspension. The absolute cell number was quantified. The splenocytes were further isolated using a mouse CD4+ T cell-specific and B cell-specific isolation kit according to the manufacturer's instructions (Miltenyi Biotec Inc.). The purity of the obtained cells was further detected using flow cytometry with CD4 or CD19 mAbs (BD-Biosciences). Only cells with a purity greater than 95% were used for further analysis.
Real-time quantitative PCR
Total RNA obtained from isolated CD4+ T and B cells was extracted using Trizol reagent (Invitrogen). Single strand cDNA was synthesized from 1 μg of total RNA using the RNA reverse transcriptase kit (Takara). Real-time quantitative PCR was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems), and the TAKARA SYBR Premix Ex Taq was used for amplification according to the manufacturer's instructions. The mRNA level of basigin was detected using the primer pair, Bsg-1: 5'-TGGACCGTGTTCACATCCAT-3' and Bsg-2: 5'-ACCTCTTTAGCATAGTAGTCCGC-3'. β-actin was detected using the primer pair: Actin-1: 5'-GGCTGTATTCCCCTCCATCG-3' and Actin-2: 5'-CCAGTTGG TAACAA TGCCATGT-3'.
Western blotting analyses
Freshly isolated CD4+ T cells and B cells were immediately homogenized in protein lysis buffer. Thirty-micrograms of protein were isolated on 10% SDS-PAGE, transferred onto a PVDF membrane, and blotted using a goat-anti-basigin antibody (R&D Systems) or rabbit-anti-β-actin antibody (R&D Systems). After several washes, the membranes were incubated with HRP-conjugated anti-goat or anti-rabbit IgG (R&D Systems) and then visualized using ECL reagents (Amersham Bioscience).
Flow cytometry
Flow cytometry was performed according to previously described methods [9]. Freshly prepared single-cell suspensions of thymuses and spleens from at least eight animals per genotype were incubated using anti-CD16/32 to block FcRγ II/III, followed by staining with fluorochrome-conjugated mAbs. All of the antibodies were purchased from BD-Biosciences. Stained cells were analyzed using a FACSCalibur flow cytometer (BD-Biosciences) and CellQuest software (BD-Biosciences). The absolute cell number of each cell subset was calculated by the percentage of stained cells × total cell number.
T cell proliferation assay
Purified CD4+ T cells (5×104 cells/well) were incubated with either medium alone or with ConA (1 μg/mL, Sigma-Aldrich), plate-bound anti-CD3 and anti-CD28 (5 μg/mL and 5 μg/mL, BD-Biosciences, respectively), and PMA/Ionomycin (25 ng/mL/500 ng/mL, Sigma-Aldrich) for 72 hours at 37°C in 96-well plates. Next, 20 μL of 20 μg/mL WST-1 (Roche) were added into each well, and the cultures were incubated at 37°C for an additional 4 hours. Next, the absorbance was measured at 450 nm and 630 nm. The proliferation index was calculated as (A450 nm - A630 nm) of the T cells cultured with different mitogens divided by (A450 nm - A630 nm) of the T cells cultured with medium alone.
Statistical analysis
The Graphpad prism software was used to manage the data. The results are presented as the means ± SD, and two-tailed Student's t-test was used to compare the significance between two groups. *P < 0.05 was considered significant.
Results
Generation of a conditional null allele for basigin in mice
The murine Bsg gene spans over 7.6 kb on chromosome 10 and contains 8 exons, including the noncoding exon 1 [31]. The full-length transcript of basigin is approximately 3.1 kb and encodes two isoforms of the basigin proteins 1 and 2, depending on the alternative splicing of exon 2. Because the gene fragment from exon 2 to exon 7 encodes over 97% of basigin amino acids, and deletion of this fragment will introduce a frame shift mutation in exon 8, a deletion of the gene fragment from exon 2 to exon 7 using Cre recombinase is expected to result in a null allele of basigin for both isoforms. To generate a conditional null allele for basigin, we flanked exon 2 and exon 7 with two loxP sites. The pfrtI vector, which contains a Neo cassette flanked by two FRT sites, a loxP site and an Hsv-tk cassette, was used to construct the conditional gene targeting vector. First, we inserted a 1.5-kb fragment located in intron 1 of basigin into the pfrtI vector for the 5' homology recombination arm, and then inserted a 3.2-kb fragment containing exons 2 to 7 downstream of the loxP site into the pfrtI vector, followed by a 3.7-kb fragment containing a second loxP site and exon 8 inserted into the pfrtI vector to form the 3' homology recombination arm (Fig. 1A). After linearization by Not I, the targeting vector was electroporated into R1 embryonic stem cells (ES). Three out of 192 (1.6%) ES clones with G418- and ganciclovir- resistance showed correct recombination as detected by Southern blot using an external 3' probe, which exhibited a 4.1-kb targeted allele (referred to as Bsgfn, fn denotes floxP-neo) band in addition to an 8.1-kb wildtype allele (referred to as Bsg+) band (Fig. 1A and 1B). The 5' arm homologous recombination event of the targeted ES clones was also confirmed by PCR using a Neo specific primer pair, P1 and P2, which amplified a unique 2.1-kb fragment of the Bsgfn allele, but not the Bsg+ allele (Fig. 1A and 1C). These results demonstrated that three targeted ES clones with the floxed basigin allele (Bsgfn) had been successfully generated.
Generation of the conditional null allele for basigin. A. Strategy for generating the conditional targeting vector, targeted Bsgfn allele, targeted Bsgfl allele and excised allele Bsg-. Flp-mediated excision of Neo cassette from the Bsgfn allele resulted in the Bsgfl allele, and Cre-mediated excision of the loxP sites resulted in the Bsg- allele. The 3' external probe for the Southern blot is indicated by the thick lines. The predicted lengths of the Southern fragments are indicated by the double arrow lines. PCR primers used to identify the targeted and deleted alleles are indicated by the arrows. N, Not I. B. Correctly targeted ES cells were confirmed by Southern blot analysis. Genomic DNA was isolated from ES cells, digested with Not I and subjected to Southern blot analysis using a 3' external probe as indicated in Fig. 1A. C. The 5' arm homologous recombination events in targeted ES cells were confirmed by PCR using primers P1 and P2. D. Deletion of the Neo cassette was identified by PCR using primers P5 and P6.
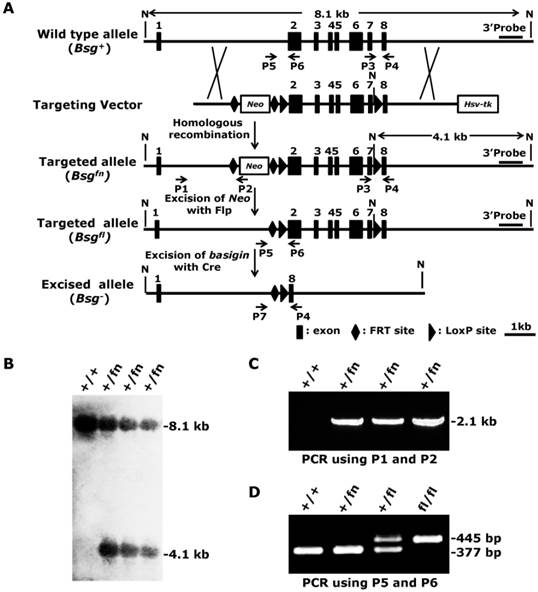
The correctly targeted ES cells were then microinjected into blastocysts obtained from 129/SV mice to generate chimeric mice according to standard protocols [32]. F1 mice were analyzed by PCR using primers P3 and P4 to validate germline transmission of the Bsgfn allele (data not shown). Because the Neo cassette located in intron 1 could potentially disrupt basigin mRNA transcription, splicing, or stability, we removed it by crossing the Bsgfn/+ mice to Flp transgenic mice [33], which resulted in Bsgfl/+ mice (Fig. 1A, f denotes floxP). The deletion of the Neo cassette was confirmed by PCR with primers P5 and P6, which amplified a 445-bp fragment from the Bsgfl allele but only a 377-bp fragment from the wildtype allele (Fig. 1A and 1D). Heterozygous Bsgfl/+ mice were intercrossed to generate homozygous Bsgfl/fl mice, which were born at the expected Mendelian ratio (Bsg+/+: Bsgfl/+: Bsgfl/fl = 54: 117: 61). Both male and female Bsgfl/fl mice were viable, fertile, and presented no genotype-dependent differences in gross morphological abnormalities, indicating that the floxed basigin allele (Bsgfl) was a functional basigin allele.
Conditional deletion of basigin allele using Lck-Cre transgenic mice
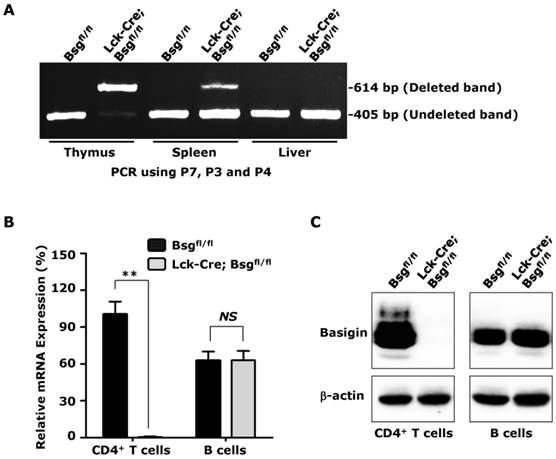
To demonstrate that the Bsgfl allele could be deleted using Cre recombinase in vivo and to obtain additional information regarding the role of basigin in thymocyte development and T lymphocyte activation, Bsgfl/fl mice were mated to Lck-Cre transgenic mice, which have been successfully used to delete floxed genes in thymocytes and T lymphocytes [34]. We designed PCR primers (Fig. 1A) to amplify the deleted allele (using primers P7 and P4; 614-bp product in length) and undeleted allele (using primers P3 and P4; 405-bp product in length). The PCR results showed no deleted band in tissues obtained from control mice, but a strong deleted band in genomic DNA from the thymus, a moderate band in the spleen, and no deleted allele band in the genomic DNA from liver of the Lck-Cre; Bsgfl/fl mice (Fig. 2A) was observed, which was consistent with the level of T lymphocytes in these tissues. However, a very weak undeleted band in the PCR results of the thymus was still present, which might have been amplified from the genomic DNA of cells lacking Cre expression in the thymus, such as thymic epithelial cells. To further investigate whether Lck-Cre-mediated excision of the Bsgfl allele was T lymphocyte-specific, we isolated CD4+ T lymphocytes and CD19+ B lymphocytes from the spleen and measured the basigin expression both at the mRNA and protein level. Real-time quantitative PCR data showed that basigin mRNA was normal in CD4+ T lymphocytes from control Bsgfl/fl mice, but was undetectable in the CD4+ T lymphocytes from Lck-Cre; Bsgfl/fl mice. However, the basigin mRNA level in B lymphocytes showed no difference between mice of the two different genotypes (Fig. 2B). Using a western blotting analysis, the specific deletion of basigin protein in CD4+ T lymphocytes of Lck-Cre; Bsgfl/fl mice was also verified using an anti-basigin antibody (Fig. 2C). These results demonstrated that the Bsgfl allele could be specifically deleted in thymocytes and subsequent T lymphocytes in Lck-Cre; Bsgfl/fl mice. In addition, the Bsgfl/fl mice could be used for tissue-specific deletion of basigin using previously developed Cre transgenic mouse models.
Deletion of the Bsgfl allele in Lck-Cre; Bsgfl/fl mice. A. PCR analysis of the basigin deletion on genomic DNA obtained from the thymus, spleen and liver. Primers P3, P4 and P7 were added in a same tube for PCR reaction in this experiment. B. Real-time quantitative PCR. The relative mRNA level of basigin in CD4+ T cells isolated from Bsgfl/fl mice was presented as 1, and the mRNA levels of basigin in other cells were expressed as a relative ratio. n = 4. **, P < 0.01. C. Western blot analysis. Total protein from the CD4+ T cells and B cells were immunoblotted using an anti-basigin antibody. β-actin was used as the loading control.
Targeted disruption of the basigin resulted in a decrease in thymic cellularity. Thymocytes were obtained from 5-week-old mice. A. Thymus weight (left) and absolute cell number of total thymocytes (right) were assessed. n = 8 per group. B. Analysis of CD4+ and CD8+ thymocytes from Lck-Cre; Bsgfl/fl mice and their control littermates using flow cytometry. C. The absolute cell number of the DN, DP, CD4SP and CD8SP thymocytes. n = 8. NS, no significance. *, P < 0.05; **, P < 0.01.

Analysis of thymocytes from Lck-Cre; Bsgfl/fl mice
General observation and physical examination apparently showed that the Lck-Cre; Bsgfl/fl mice showed no obvious difference in development and growth compared to the control mice (Bsgfl/fl). No defects were found in the size and tissue structure of major organs in adult mice, including heart, lung, liver, kidney, and other organs. Previous reports have shown the involvement of basigin in thymus development ex vivo; thus, we examined whether the development of thymus was impaired in the absence of basigin in Lck-Cre; Bsgfl/fl mice. We isolated the thymus from 5-week-old mice and found a significant reduction in thymus weight in Lck-Cre; Bsgfl/fl mice compared to that in control mice (Fig. 3A left). The total number of thymocytes in Lck-Cre; Bsgfl/fl mice was approximately 29% lower compared to control mice (Fig. 3A right). Quantification of individual thymocyte subsets using flow cytometry with CD4 and CD8 mAbs showed that the number of DP and CD4SP thymocytes from Lck-Cre; Bsgfl/fl mice was reduced by 32% and 22%, respectively, compared to control mice (Fig. 3B and 3C). However, the absolute number of DN (double-negative, CD4-CD8-) and CD8SP (CD4-CD8+) thymocytes showed no significant difference between mice of the two different genotypes (Fig. 3C). Similar results were also found in the thymus from 2-week-old Lck-Cre; Bsgfl/fl mice and their control mice (Supplementary Material: Fig. S1). The above results indicated that basigin expression in T lymphocytes is necessary for thymocyte development, particularly at the DP and CD4SP stage. Since the observed drop of cell number in thymus mainly came from DP subset, we assessed freshly isolated thymocytes by Annexin V versus PI staining to investigate whether increased apoptosis of DP thymocytes accounted for the reduced thymocyte numbers. However, there were no significant difference of apoptotic DP thymocytes between Lck-Cre; Bsgfl/fl mice and the control mice (Supplementary Material: Fig. S2).
Analysis of peripheral T cells from Lck-Cre; Bsgfl/fl mice
We also analyzed the number of peripheral T cells isolated from the Lck-Cre; Bsgfl/fl mice. In contrast to the population variation found in thymocytes, the total number of splenocytes was slightly, but not significantly, reduced in Lck-Cre; Bsgfl/fl mice compared to control mice (Fig. 4A). An analysis of splenocytes using CD4 and CD8 mAbs also did not show significant changes in the percentage and total number of CD4+ T lymphocytes and CD8+ T lymphocytes (Fig. 4B and 4C). Because basigin has previously been reported to be highly expressed on activated T lymphocytes, we next analyzed whether T lymphocyte activation was affected in the absence of basigin expression. CD4+ T lymphocytes were isolated from the spleen, and stimulated with ConA, anti-CD3/CD28 antibodies, PMA/Ionomycin and control medium. In the control medium treatment group, no difference in the proliferative index was observed between the Lck-Cre; Bsgfl/fl mice and control mice. However, after stimulation with ConA and anti-CD3/CD28 antibodies, the proliferation index of the CD4+ T lymphocytes isolated from Lck-Cre; Bsgfl/fl mice was significantly increased compared to that of the CD4+ T lymphocytes from the control mice (Fig. 4D). In contrast, the proliferation index of CD4+ T lymphocytes from Lck-Cre; Bsgfl/fl mice stimulated with PMA/Ionomycin was the same as that of the control CD4+ T lymphocytes. To examine whether deletion of basigin enhanced T cell activation via T cell receptor (TCR), we measured the expression of CD69 on CD4+ T cells stimulated with anti-CD3/CD28 antibodies. As shown in Figure 4E, expression of CD69 was nearly undetectable in unstimulated CD4+ T cells from both Lck-Cre; Bsgfl/fl mice and control mice. However, expression level of CD69 on the CD4+ T cells from Lck-Cre; Bsgfl/fl mice was higher than that from control mice after stimulation. Consistent with the proliferation index result, the percentages of CD69+ cells were also significantly increased in stimulated CD4+ T cells from Lck-Cre; Bsgfl/fl mice compared to that from control mice (Figure 4F). Taken together, the above results indicated that basigin may regulate T cell activation via T cell receptor activation and may not directly regulate Ca2+ storage release and protein kinase C signaling pathway.
Analysis of peripheral T cells from Lck-Cre; Bsgfl/fl mice and their control mice. T lymphocytes were freshly isolated from the spleens of 5-week-old Lck-Cre; Bsgfl/fl mice and their control mice. A. Analysis of the spleen weight and absolute cell number of total splenocytes. n = 8. B. Analysis of CD4+ and CD8+ T lymphocytes using flow cytometry. C. The absolute cell number of CD4+ and CD8+ T lymphocytes. n = 8. D. Proliferation analysis of CD4+ T lymphocytes stimulated with medium, ConA, anti-CD3/CD28 antibodies and PMA/Ionomycin. n = 4. E. CD69 expression in CD4+ T lymphocytes stimulated or unstimulated with anti-CD3/CD28 antibodies for 18 hours. F. Percentages of CD69 positive cell in CD4+ T lymphocytes. n = 3. *, P < 0.05. NS, no significance.

Discussion
In this study, we generated a conditional null allele for basigin (Bsgfl) in mice using the Cre-loxP system. Bsgfl/fl mice were born at the expected Mendelian ratio and exhibited a growth rate similar to wildtype mice. Next, we generated Lck-Cre; Bsgfl/fl mice by intercrossing the Bsgfl/fl mice with Lck-Cre transgenic mice. In Lck-Cre; Bsgfl/fl mice, the floxed basigin gene was specifically inactivated in thymocytes and T lymphocytes but not in B lymphocytes or other somatic cells. A phenotypic analysis revealed that Lck-Cre; Bsgfl/fl mice showed no obvious difference in birth and growth rates compared to control mice, except for significant reduction in thymus weight and total number of thymocytes. Inactivation of basigin in T lymphocytes also significantly decreased the number of DP and CD4SP cells in the adult thymus, and enhanced the proliferation of CD4+ T cells upon ConA or anti-CD3/CD28 stimulation.
Basigin is a multifunctional protein that is thought to be involved in a variety of physiological and pathological activities, such as embryo development, reproduction, immune response and tumor metastasis [1, 3-5, 28]. However, the in vivo functions of basigin are still unclear, partly due to the shortage of appropriate experimental animal models. In addition, basigin knockout mice have been previously generated using a conventional gene targeting strategy [3]; however, only a very small number of Bsg-/- embryos survive to birth and develop into small, weak, and infertile adult mice. These developmental challenges prevented an elaborate phenotypic analysis and the establishment of some disease models. To overcome these disadvantages, Kadomatsu et al. generated a hybrid Bsg-/- mouse line known as “reverse F2,” by intercrossing F2 animals from the F1 hybrid offspring of 129/SV chimera and C57BL/6J animals with a C57BL/6J congenic mouse line [35]. The “reverse F2” Bsg-/- mice were born at an expected frequency (~20%), possibly because of the specific genetic background and the composition of the flanking region of the Bsg gene, but not the normal function of basigin. Moreover, because basigin is expressed in different cell types and may function via cell-cell interaction, knocking out basigin expression in all cells prevented the further study of its role in specific cell types and its corresponding cellular and molecular mechanisms. Here, we generated a conditional null allele for basigin using the Cre-loxP system. In contrast to the previously reported Bsg-/- mice, Bsgfl/fl mice showed no fertility and growth defects, indicating that the insertion of the loxP sites had no effect on basigin gene activity. Intercrossing Bsgfl/fl mice with Lck-Cre transgenic mice demonstrated an effective knockout of the basigin gene in cells with lck promoter activity (thymocytes and T lymphocytes), which demonstrated that Bsgfl/fl mice could be a very useful tool to study basigin's in vivo physiological and pathological functions in specific cell types and at different development stages.
Lck-Cre; Bsgfl/fl mice also showed no difference in birth and growth rate compared to control mice, which enabled further analysis of basigin's function in thymus development. First, we found that the thymuses of Lck-Cre; Bsgfl/fl mice were significantly smaller compared to control Bsgfl/fl mice, which was accompanied by a significant reduction in the number of thymocytes (Fig. 3A). Reno et al. previously reported that a basigin-specific mAb (RL73.2) could block the ex vivo development of fetal thymocytes into mature T cells by reducing the number of live cells in a fetal thymic organ culture, which predominantly occurred at the transition between the DN3 and DN4 stages; however, treatment with the mAb did not result in an increase in cell death in adult thymocytes [9]. Because the expression of the Lck-Cre transgene used in our study might be initiated after the DN2 stage [34], the inactivation of basigin was expected to begin as early as the DN3 stage. This may have contributed to the weak undeleted band observed in the PCR test using thymus genomic DNA obtained from the Lck-Cre; Bsgfl/fl mice (Fig. 2A). However, Lck-Cre; Bsgfl/fl mice did not show any significant change in the number of DN cells in the adult thymus, and exhibited a significantly decreased cell number of DP and CD4SP cells in the adult thymus compared to controls, which were inconsistent with Reno's findings. Our results showed an important in-vivo function of basigin in thymus development at the DP stage and CD4SP stage.
In contrast to the population variation found in the thymus, the relative proportions and absolute numbers of peripheral CD4 and CD8 T cell were similar between Lck-Cre; Bsgfl/fl mice and controls (Fig. 4B and 4C). The possible reason was that maintenance of the peripheral T cell population does not only depend on the influx of thymic emigrants cells, but also depend on the homeostatic mechanism [36]. When the number of peripheral T cells is severely depleted in some pathological case, the remaining T cells sense the increased availability of “space” and undergo spontaneous proliferation to re-establish homeostasis. Although the molecular basis remains unclear, interaction of TCR with self-peptide/MHC molecules seem to participate in the homeostatic proliferation of peripheral T cells [37]. We found that CD4+ T cells from Lck-Cre; Bsgfl/fl mice expressed higher level of CD69 (Fig 4E) and proliferated stronger after stimulation with anti-CD3/CD28 antibodies (Fig 4D and 4F), compared with the control cells. These results suggested that deletion of basigin may plays an important role in T cell activation and proliferation via elevating TCR signal strength. It is possible that deletion of basigin also augments the signals delivered by the complex of TCR and self-peptide/MHC molecules, and facilitates homeostatic proliferation of CD4+ T cells in Lck-Cre; Bsgfl/fl mice.
Igakura et al. previously found that the mitogenic response of lymphocytes upon a mixed lymphocyte reaction was greater in Bsg-/- mice [3, 29]. Our previous in vitro study showed that the engagement of basigin with a specific anti-basigin mAb in human T lymphocytes strongly inhibited T cell proliferation and disrupted the formation of the immunological synapse [12]. To the best of our knowledge, we are the first to show that the genetic ablation of basigin in T lymphocytes alone did not affect the proliferation or survival of CD4+ T cells isolated from the adult spleen but, rather, resulted in a significantly enhanced proliferation of CD4+ T cells upon ConA or anti-CD3/CD28 stimulation. This finding suggested that basigin may not to be a general regulator of T cell survival or proliferation but may suppress the activation or proliferation of T cells via TCR activation. Cytosolic Ca2+ dynamics is important for the precise control of T cell activation, and relies on the extracellular Ca2+ influx following TCR activation and intracellular storage release evoked by downstream signals of TCR stimulation. Our previous study showed that anti-basigin mAb could inhibit cytosolic Ca2+ concentrations when added together with anti-CD3/CD28 antibodies. However, in the present study, no difference in the T cell proliferation index was observed between the Lck-Cre; Bsgfl/fl mice and control mice upon PMA/Ionomycin stimulation, suggesting that the suppression of basigin on Ca2+ mobility may rely on TCR activation, or that basigin may only inhibit the TCR-mediated Ca2+ influx, but not the release of intracellular Ca2+. Further elucidation of the mechanistic functions of basigin in thymus development and T lymphocyte function are required and are currently ongoing in our lab. Moreover, Lck-Cre; Bsgfl/fl mice will be a useful tool for the exploration of basigin function in T lymphocytes.
To the best of our knowledge, this study presented the first in vivo evidence for the function of basigin in thymus development. In addition, the successful generation of the conditional null allele for basigin provides a useful tool for the tissue- and developmental stage-specific ablation of basigin using previously developed Cre transgenic mouse models.
Supplementary Material
Fig.S1 - Fig.S2.
Acknowledgements
We thank Dr. Hua Han for providing us with Lck-Cre transgenic mice. This work is supported by China National Basic Research Program (NO. 2009CB521704); China National Key Project of Scientific and Technical Supporting Programs (NO. 2006BAI23B02); Key Project of China National Natural Science Fund (NO. 81030058); China National Major Project (NO. 2013ZX09301301); the National Science and Technology Major Projects (NO. 2012AA020806) and the China National Natural Science Foundation (NO. 30800573).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Muramatsu T, Miyauchi T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol Histopathol. 2003;18:981-7
2. Biswas C, Zhang Y, Decastro R. et al. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 1995;55:434-9
3. Igakura T, Kadomatsu K, Kaname T. et al. A null mutation in basigin, an immunoglobulin superfamily member, indicates its important roles in peri-implantation development and spermatogenesis. Dev Biol. 1998;194:152-65
4. Toyama Y, Maekawa M, Kadomatsu K. et al. Histological characterization of defective spermatogenesis in mice lacking the basigin gene. Anat Histol Embryol. 1999;28:205-13
5. Kuno N, Kadomatsu K, Fan QW. et al. Female sterility in mice lacking the basigin gene, which encodes a transmembrane glycoprotein belonging to the immunoglobulin superfamily. FEBS Lett. 1998;425:191-4
6. Fan QW, Yuasa S, Kuno N. et al. Expression of basigin, a member of the immunoglobulin superfamily, in the mouse central nervous system. Neurosci Res. 1998;30:53-63
7. Naruhashi K, Kadomatsu K, Igakura T. et al. Abnormalities of sensory and memory functions in mice lacking Bsg gene. Biochem Biophys Res Commun. 1997;236:733-7
8. Philp NJ, Ochrietor JD, Rudoy C. et al. Loss of MCT1, MCT3, and MCT4 expression in the retinal pigment epithelium and neural retina of the 5A11/basigin-null mouse. Invest Ophthalmol Vis Sci. 2003;44:1305-11
9. Renno T, Wilson A, Dunkel C. et al. A role for CD147 in thymic development. J Immunol. 2002;168:4946-50
10. Kasinrerk W, Fiebiger E, Stefanova I. et al. Human leukocyte activation antigen M6, a member of the Ig superfamily, is the species homologue of rat OX-47, mouse basigin, and chicken HT7 molecule. J Immunol. 1992;149:847-54
11. Staffler G, Szekeres A, Schutz GJ. et al. Selective inhibition of T cell activation via CD147 through novel modulation of lipid rafts. J Immunol. 2003;171:1707-14
12. Hu J, Dang N, Yao H. et al. Involvement of HAb18G/CD147 in T cell activation and immunological synapse formation. J Cell Mol Med. 2010;14:2132-43
13. Damsker JM, Okwumabua I, Pushkarsky T. et al. Targeting the chemotactic function of CD147 reduces collagen-induced arthritis. Immunology. 2009;126:55-62
14. Zhu P, Lu N, Shi ZG. et al. CD147 overexpression on synoviocytes in rheumatoid arthritis enhances matrix metalloproteinase production and invasiveness of synoviocytes. Arthritis Res Ther. 2006;8:R44
15. Gabison EE, Hoang XT, Mauviel A. et al. EMMPRIN/CD147, an MMP modulator in cancer, development and tissue repair. Biochimie. 2005;87:361-8
16. Boulos S, Meloni BP, Arthur PG. et al. Evidence that intracellular cyclophilin A and cyclophilin A/CD147 receptor-mediated ERK1/2 signalling can protect neurons against in vitro oxidative and ischemic injury. Neurobiol Dis. 2007;25:54-64
17. Seko Y, Fujimura T, Taka H. et al. Hypoxia followed by reoxygenation induces secretion of cyclophilin A from cultured rat cardiac myocytes. Biochem Biophys Res Commun. 2004;317:162-8
18. Zhou S, Zhou H, Walian PJ. et al. CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer's disease amyloid beta-peptide production. Proc Natl Acad Sci U S A. 2005;102:7499-504
19. Xu J, Xu HY, Zhang Q. et al. HAb18G/CD147 functions in invasion and metastasis of hepatocellular carcinoma. Mol Cancer Res. 2007;5:605-14
20. Tang Y, Nakada MT, Kesavan P. et al. Extracellular matrix metalloproteinase inducer stimulates tumor angiogenesis by elevating vascular endothelial cell growth factor and matrix metalloproteinases. Cancer Res. 2005;65:3193-9
21. Tang Y, Nakada MT, Rafferty P. et al. Regulation of vascular endothelial growth factor expression by EMMPRIN via the PI3K-Akt signaling pathway. Mol Cancer Res. 2006;4:371-7
22. Dai JY, Dou KF, Wang CH. et al. The interaction of HAb18G/CD147 with integrin alpha6beta1 and its implications for the invasion potential of human hepatoma cells. BMC Cancer. 2009;9:337
23. Yang JM, O'Neill P, Jin W. et al. Extracellular matrix metalloproteinase inducer (CD147) confers resistance of breast cancer cells to Anoikis through inhibition of Bim. J Biol Chem. 2006;281:9719-27
24. Fanelli A, Grollman EF, Wang D. et al. MCT1 and its accessory protein CD147 are differentially regulated by TSH in rat thyroid cells. Am J Physiol Endocrinol Metab. 2003;285:E1223-9
25. Ishibashi Y, Matsumoto T, Niwa M. et al. CD147 and matrix metalloproteinase-2 protein expression as significant prognostic factors in esophageal squamous cell carcinoma. Cancer. 2004;101:1994-2000
26. Reimers N, Zafrakas K, Assmann V. et al. Expression of extracellular matrix metalloproteases inducer on micrometastatic and primary mammary carcinoma cells. Clin Cancer Res. 2004;10:3422-8
27. Sienel W, Polzer B, Elshawi K. et al. Cellular localization of EMMPRIN predicts prognosis of patients with operable lung adenocarcinoma independent from MMP-2 and MMP-9. Mod Pathol. 2008;21:1130-8
28. Li Y, Xu J, Chen L. et al. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology. 2009;54:677-87
29. Igakura T, Kadomatsu K, Taguchi O. et al. Roles of basigin, a member of the immunoglobulin superfamily, in behavior as to an irritating odor, lymphocyte response, and blood-brain barrier. Biochem Biophys Res Commun. 1996;224:33-6
30. Wu X, Ding H. Generation of conditional knockout alleles for PDGF-C. Genesis. 2007;45:653-7
31. Miyauchi T, Jimma F, Igakura T. et al. Structure of the mouse basigin gene, a unique member of the immunoglobulin superfamily. J Biochem. 1995;118:717-24
32. Kühn R, Wurst W. Gene knockout protocols. 2nd ed. New York, USA: Humana Press. 2009
33. Rodriguez CI, Buchholz F, Galloway J. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139-40
34. Fu T, Zhang P, Feng L. et al. Accelerated acute allograft rejection accompanied by enhanced T-cell proliferation and attenuated Treg function in RBP-J deficient mice. Mol Immunol. 2011;48:751-9
35. Chen S, Kadomatsu K, Kondo M. et al. Effects of flanking genes on the phenotypes of mice deficient in basigin/CD147. Biochem Biophys Res Commun. 2004;324:147-53
36. Surh CD, Sprent J. Regulation of mature T cell homeostasis. Semin Immunol. 2005;17:183-91
37. Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848-62
Author contact
![]() Corresponding authors: Zhi-Nan Chen, Cell Engineering Research Centre & Department of Cell Biology, State Key Laboratory of Cancer Biology, Fourth Military Medical University, 169 West Changle Street, Xi'an 710032, China. Tel: +86-29-84773243; Fax: +86-29-83293906; E-mail: znchenedu.cn. Xiao Yang, Genetic Laboratory of Development and Disease, Institute of Biotechnology, 20 Dongdajie, Fengtai District, Beijing 100071, China. Tel: +86-10-63895937; Fax: +86-10-63895937; E-mail: yangxbmi.ac.cn.
Corresponding authors: Zhi-Nan Chen, Cell Engineering Research Centre & Department of Cell Biology, State Key Laboratory of Cancer Biology, Fourth Military Medical University, 169 West Changle Street, Xi'an 710032, China. Tel: +86-29-84773243; Fax: +86-29-83293906; E-mail: znchenedu.cn. Xiao Yang, Genetic Laboratory of Development and Disease, Institute of Biotechnology, 20 Dongdajie, Fengtai District, Beijing 100071, China. Tel: +86-10-63895937; Fax: +86-10-63895937; E-mail: yangxbmi.ac.cn.

 Global reach, higher impact
Global reach, higher impact