10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):883-891. doi:10.7150/ijbs.24616 This issue Cite
Research Paper
iRSpot-Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC
1. Key Laboratory for Neuro-Information of Ministry of Education, School of Life Science and Technology, Center for Informational Biology, University of Electronic Science and Technology of China, Chengdu 610054, China;
2. Computer Department, Jingdezhen Ceramic Institute, Jingdezhen, 333403, China;
3. School of Life Science and Technology, Inner Mongolia University of Science and Technology, Baotou, 014010, China.
4. Department of Physics, School of Sciences, and Center for Genomics and Computational Biology, North China University of Science and Technology, Tangshan 063000, China
5. Gordon Life Science Institute, Boston, MA 02478, USA
Abstract
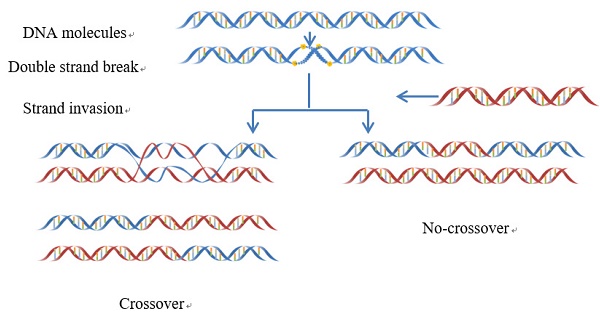
Meiotic recombination caused by meiotic double-strand DNA breaks. In some regions the frequency of DNA recombination is relatively higher, while in other regions the frequency is lower: the former is usually called “recombination hotspot”, while the latter the “recombination coldspot”. Information of the hot and cold spots may provide important clues for understanding the mechanism of genome revolution. Therefore, it is important to accurately predict these spots. In this study, we rebuilt the benchmark dataset by unifying its samples with a same length (131 bp). Based on such a foundation and using SVM (Support Vector Machine) classifier, a new predictor called “iRSpot-Pse6NC” was developed by incorporating the key hexamer features into the general PseKNC (Pseudo K-tuple Nucleotide Composition) via the binomial distribution approach. It has been observed via rigorous cross-validations that the proposed predictor is superior to its counterparts in overall accuracy, stability, sensitivity and specificity. For the convenience of most experimental scientists, the web-server for iRSpot-Pse6NC has been established at
Keywords: Recombination spot, 5-step rules, Key hexamers, PseKNC, SVM, Webserver

 Global reach, higher impact
Global reach, higher impact