Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(9):1163-1171. doi:10.7150/ijbs.20792 This issue Cite
Review
Fibroblast Growth Factor Receptor 2 Signaling in Breast Cancer
Faculty of Health Sciences, University of Macau, Macau SAR, China
Received 2017-4-29; Accepted 2017-5-18; Published 2017-9-5
Abstract
Fibroblast growth factor receptor 2 (FGFR2) is a membrane-spanning tyrosine kinase that mediates signaling for FGFs. Recent studies detected various point mutations of FGFR2 in multiple types of cancers, including breast cancer, lung cancer, gastric cancer, uterine cancer and ovarian cancer, yet the casual relationship between these mutations and tumorigenesis is unclear. Here we will discuss possible interactions between FGFR2 signaling and several major pathways through which the aberrantly activated FGFR2 signaling may result in breast cancer development. We will also discuss some recent developments in the discovery and application of therapies and strategies for breast cancers by inhibiting FGFR2 activities.
Keywords: Breast cancer, FGFR2, mutation, polymorphism, inhibitor.
Introduction
Fibroblast growth factor receptor 2 (FGFR2) belongs to a family of four typical membrane-bound receptor tyrosine kinases (RTKs) [1-4]. Although an additional FGF receptor, FGFR5, has been identified, it does not contain the intracellular tyrosine kinase domain [5]. FGFRs mediate signaling from fibroblast growth factors (FGFs), which constitute of a gene family of at least 22 members and have numerous important functions, including developmental induction, pattern formation, cell growth and differentiation, as well as survival and death [1-4]. It has been shown that mutations of FGFRs were able to facilitate tumor growth by driving cell proliferation and survival [6], but also could suppress tumor growth [7]. Consistently, studies of mouse models have revealed that FGFR2 not only serves as an oncogenic gene [8, 9], but also acts as a tumor suppressor in a certain intracellular environment [10-12].
In this review, we discuss insights into the mechanism of FGFR2 regulation that has emerged from structural and functional studies. We will review the integrated networks that result from the interplay among the complex signaling pathways activated by FGFR2 during breast cancer formation. Finally, we will explore the applications of therapies and strategies for breast cancers driven by FGFRs activities in clinical conditions.
Structure of FGFR2 and its signaling pathways
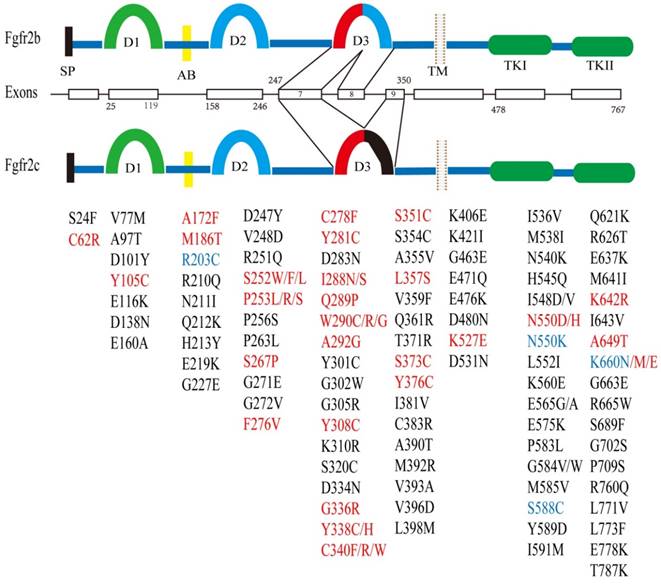
The FGFR2 gene is located at chromosome 10q26 and contains 20 exons in humans and 19 exons in the mouse. FGFR2 encodes two major isoforms through alternative splicing, FGFR2b and FGFR2c, which have distinct function domains that specifically recognize a variety of fibroblast growth factors (FGFs) [1, 13-16]. As a membrane bound receptor, FGFR2 contains an extracellular region, which is made up of signal peptides (SP), three immunoglobulin domains (D1, D2, and D3) and an acid box (AB) [17]. This is followed by a cross-membrane domain, a split tyrosine kinase (TK) domain (including TK1 and TK2) and a short carboxyl-terminal tail (Figure 1).
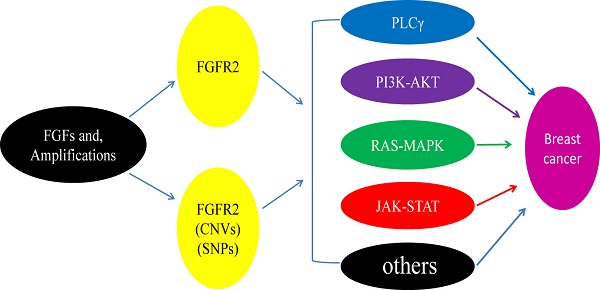
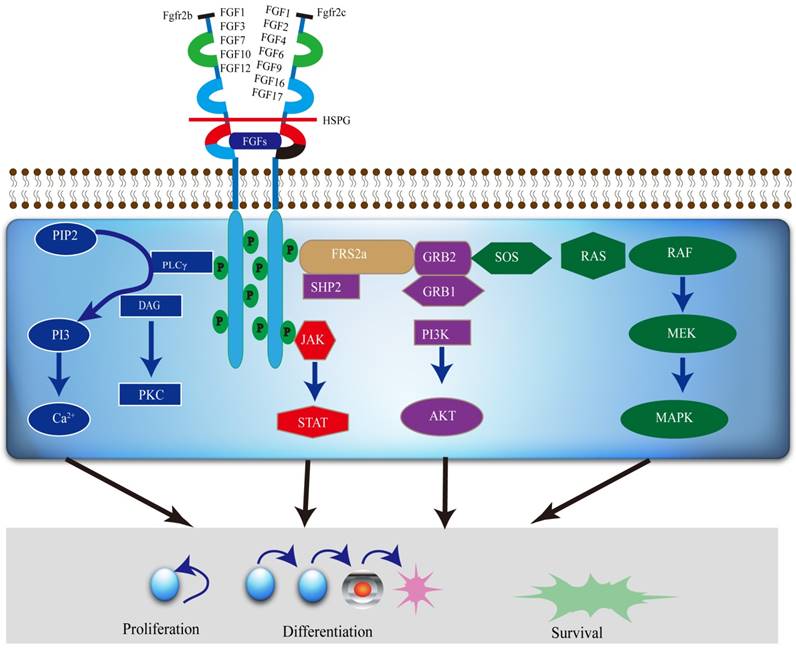
The FGFR2 signaling pathways divide into two streams. One is dependent on FGFR substrate 2α (FRS2α) [18], while the other is not. It is involved in major four pathways, including RAS-MAPK (mitogen-activated protein kinase), PLCγ (Phospholipase C-γ), PI3K (phosphoinositide 3-kinase), and Janus kinase/signal transducer and activator of transcription (JAK/STAT) (Figure 2). FGFR2 acts through multiple downstream signaling pathways that play vital roles in cell proliferation [12, 19, 20], survival [21], differentiation [22] and drug resistance [23]. FGFR substrate 2 (FRS2) is a key adaptor protein that binds to the juxtamembrane region of FGFR2 through its phosphotyrosine-binding (PTb) domains. The activated FGFR2 phosphorylates FRS2 on several sites, allowing the recruitment of the adaptor proteins 'Son of Sevenless' (SOS) and growth factor receptor bound 2 (GRb2) to activate RAS and the downstream MAPK pathways. A separate complex involving GRb2 associated binding protein 1 (GAb1) recruits a complex, which includes PI3K, and this activates a PI3K-AKT signaling pathway. Another FGFR2 binding partner is phospholipase Cγ (PLCγ), which binds at the carboxyl‑terminal tail on auto-phosphorylation of FGFR2. After PLCγ is activated, it hydrolyses phosphatidylinositol 4,5 biphosphate (PIP2) to phosphatidylinositol 3,4,5 triphosphate (PIP3) and diacylglycerol (DAG), activating protein kinase C (PKC), which enhance the stimulation of the MAPK pathway by phosphorylating RAF. Several other pathways are also activated by FGFR2, such as the JAK-STAT pathway [24] (Figure 2).
Amplification of FGFR2 in breast cancer
Breast cancer occurs in women worldwide and is the causes of the highest cancer lethality in females of developing countries [25]. Currently about 1.7 million cases of breast cancers and about half million deaths are reported annually in 2012 [26]. With increasing incidence and mortality, breast cancer will certainly be a big public problem not only in developing countries, but also in developed countries.
Structure of the FGFR2 gene and its somatic mutations with their relative locations. There are two major isoforms of FGFR2, i.e. FGFR2b (upper), and FGFR2c (lower) that are caused by alternative splicing of exons 8 and 9. Both isoforms contain multiple functional domains as indicated. SP: signal peptide; D1-3: immunoglobulin I-III; AB: acid box; TM: transmembrane; TK: tyrosine kinase. Somatic mutations in FGFR2 identified in development syndrome and cancers are presented in red font, and in breast cancer is highlighted in blue font. Mutations present only in cancer are shown in black font. The residue numbers are according to Table 3.
A diagram of the FGFR2 mediated signaling pathways. Various FGFs bind to FGFR2b or FGFR2c, respectively, with HSPG as a cofactor and induce the formation of ternary FGFs-FGFR2-HS complex, which activates the FGFR2 intracellular tyrosine kinase domain by phosphorylation of specific tyrosine residues. The activated TK of FGFR2, in turn, triggers activation of multiple downstream signaling pathways, which not only regulate differentiation and gene expression in the nucleus, but also drive cell proliferation and survival as well.
Multiple genetic aberrations in FGFR2 triggering the activation of up and/or downstream FGFR2 signaling pathways have been identified in breast cancer. For instance, 6 out of 165 (3.6%) of triple-negative breast cancers (TNBC) has been found to carry an amplification of FGFR2 (10q26) [27]. Researchers also screened 51 breast cancer cell lines and found two of them, MFM223 and SUM52PE, carry FGFR2 genes amplification and protein overexpression. Both cell lines were confirmed to be (TNBC) cell lines, and the amplification of FGFR2 results in the activation of PI3K-AKT signaling, leading to the inhibition of apoptosis [27]. FGFR2 gene amplification is not only found in breast cancer cell lines, but also in normal breast and tumor tissues [28-30]. In addition, Sun et al. (2011) reported that 64.8% (81/125) and 56.8% (71/125) of breast cancers expressed FGFR2 in cytoplasm and the nucleus, respectively. They showed that FGFR2 expression in cytoplasmic was significantly associated with tumor size and the tumor, node and metastasis (TNM) stage, and higher levels of FGFR2 in the cytoplasm and nucleus were associated with much lower overall survival and disease-free survival rates than lower levels of FGFR2 [31]. However, Lee et al. (2014) reported that FGFR2 amplification did not affect patient survival [32].
In vitro studies showed that FGFR2 activates ERK directly and binds to phosphorylation POU1F1 at thr75, which inhibits transcription of the double-strand break repair protein Mre11A (Mre11) via interaction with the Mre11 promoter, and promotes breast cancer formation [33]. Wei et al. (2015) expressed FGFR2 in MCF-7 cells and found that FGFR2 activates HER2 through FGF7/FGFR2-mediated ADAM10 upregulation, which results in the enhanced AKT, STAT3, ERK1/2 signaling leading to the activation of HER2 [34, 35]. Ezzat et al. (2012) found that FGFR2 regulates epithelial cell-stromal cell communications through tissue-specific FGF signals in cancer progression. In addition, their data also indicated that FGFR2 isoforms negatively regulate NF-kB nuclear translocation and activity, and reduce the growth of MDA-MB-231 tumor cells [36]. FGFR2 was also found to facilitate the development of breast cancer by promoting cell self-renewal through interacting with NF-kB signals [37].
Single Nucleotide Polymorphisms (SNPs) in intron 2 of FGFR2
| SNPs ID | Population | Number Case /Control | OR(95%CI) | Ref. |
|---|---|---|---|---|
| rs35054928 | Mix | 93010/107391 | 1.05 (0.93-1.18) | [89-91] |
| rs45631563 | 0.80 (0.76-0.85) | |||
| rs11200014 | 1.10 (0.84-1.21) | |||
| rs2981579 | 1.20 (1.11-1.29) | |||
| rs2303568 | China | 2073/2084 | 0.94 (0.78-1.13) | [92] |
| rs755793 | 1.00 (0.80-1.12) | |||
| rs3135730 | 0.93 (0.78-1.12) | |||
| rs1078806 | 0.94 (0.75-1.22) | |||
| rs2981582 | Mix | 93010/107391 | 1.22 (1.18-1.27) | [51, 93-97] |
| rs7895676 | 1.22 (0.97-1.48) | |||
| rs1219648 | 1.23 (1.19-1.26) | |||
| rs2420946 | 1.23 (1.18-1.29) | |||
| rs2981578 | 0.81 (0.79-0.83) | |||
| rs1078806 | 1.20 (1.04-1.38) | |||
| rs3750817 | 0.85 (0.79-0.91) | |||
| rs3135718 | 1.27(1.14-1.41) | |||
| rs2981575 | European | 1187/1193 | 0.88 (0.78-0.99) | [98] |
| rs1078806 | Sardinian | 1698/2178 | 0.77 (0.69-0.86) | [99] |
| rs2860197 | 0.76 (0.68-0.86) | |||
| rs2912774 | 0.77 (0.68-0.86) | |||
| rs2912780 | 1.30 (1.16-1.46) | |||
| rs2936870 | 0.77 (0.68-0.86) | |||
| rs3135774 | American | 3663/4687 | 1.47 (1.18-1.83) | [61] |
| rs10736303 | North Carolina | 1247/1105 | 1.25 (1.18-1.32) | [88, 100] |
| rs2162540 | 1.31 (1.15-1.48) | |||
| rs1896395 | Europe | 1972/1776 | 0.87 (0.69-1.10) | [101] |
| rs17102287 | China | 388/428 | 1.08 (0.88 -1.34) | [102] |
| rs17542768 | 1.28 (0.88-1.87) | |||
| rs10510097 | 0.86 (0.68-1.07) |
Note: Mix—China, Europe, Japan, American, Taiwanese, et al.
A further relationship between the FGFR2 signaling pathway and breast cancer has been illustrated by using GWAS (Genome-wide association study) [38, 39]. Analyzing 4,398 (women with breast cancer) familial breast cancer causes and 4,316 controls, five single nucleotide polymorphisms (SNPs) (rs7895676, rs2912781, rs10736303, rs2912778 and rs2981582) and in the non-coding region of FGFR2 were found to have a significant association with breast cancer [38]. Meanwhile, Hunter et al. (2007) also identified four other SNPs, rs11200014, rs2420946, rs1219648 and rs2981579, in intron 2 of FGFR2 that are associated with the risk of breast cancer [39]. So far, a much greater breast cancer risk associated SNPs have been discovered in intron 2 of FGFR2 (Table 1). FGFR2 polymorphisms are a risk factor associated with increased breast cancer (BC) susceptibility, these associations vary in different ethnic populations, such as China, Japan, Indian, Pakistani African American, Europe and so on, yet how they enhance breast cancer risk remains elusive.
Breast cancer associated gene 1 and 2 (BRCA1 and BRCA2) are related to a family history of breast cancer. Women having germline mutations in BRCA1 have a 50-80% risk of developing breast cancer by 70 years of age [40-45]. In BRCA1-associated tumors showed increased expression of the FGFR2 gene [46], particularly in ER-α positive breast cancer [47]. SNPs in the FGFR2 gene were associated with a significantly increased risk of luminal A, luminal B (ER/PR+, HER2+), or HER2+/ER- disease, but none were associated with basal-like (ER-, PR-, HER-) disease [48]. In BRCA1 carriers, rs2981582 of FGFR2 showed significant differences with a strong association for ER-positive or PR-positive disease but not ER-negative or PR-negative disease. There were no differential associations between ER states and PR status in BRCA2 mutation carriers [49, 50]. Interestingly, the GA and AA genotypes of FGFR2 rs2981582 are associated with a reduced the rate of breast cancer in Heilongjiang Province in Northeast China [51], whereas the AA genotypes of FGFR2 rs2981582 had an increased breast cancer risk in the Swedish population [52]. These contradictory observations suggest that gene-gene or gene-environmental interaction regulates the corporation between SNPs and the risk of breast cancer in difference regions or ethnics. Furthermore, SNPs (rs1078806, rs2420946, rs2981579, rs2981582) in intron 2 of FGFR2 is more highly linked to ER-positive and PR-Positive than ER-negative cancers in European and Asian [53, 54]. However, rs1219648 exhibited no differences associated between ER-positive cancer, ER-negative cancer and TNBC [55]. Amusingly, Andersen, et al. (2014) using 869 postmenopausal breast cancer cases and 808 postmenopausal community controls to evaluate rs1219648 for the association risk of breast cancer and found FGFR2 rs1219648 is more strongly associated with risk in estrogen-only hormone users [56]. Meanwhile, in a Chilean population, rs2981582, rs2420946 and rs1219648 are more significantly linked to familial breast cancer and early-onset non-familial breast cancer [57, 58]. One study suggested that rs2981582 and rs1219648 not only interacts with transcription factors Oct-1/Runx2, but C/EBPβ binding sites as well. Further study suggests that FGFR2 acts through ER and OCT1 activates C/EBPβ transcription [59], which may be a reason why the risk of rs2981582 is strongly associated with ER-positive but not ER-negative breast cancer [60]. Indeed, further analysis revealed that rs2981582 is associated with all breast cancer tumor types except ER-/PR- tumors [61, 62]. It was also shown that Histone3/4 acetylation is involved in downstream splicing sites of FGFR2 in breast cancer, and rs2981578 harbors a potential Oct/Runx2 binding site displaying H4 deacetylation [63]. These transcription factors are occupied in vivo and lead to increased FGFR2 expression. Moreover, Murillo et al. (2013) suggested that rs2981582 is strongly associated with breast cancer not only in alcohol consumption in Mexican women [64], but also in physical activity (swimming, running, and playing basketball, etc.) in Guangzhou as well [65]. Studying 457 male breast cancer cases and 1073 healthy male and female controls, Nick et al. (2011) found rs2981579 in FGFR2 increases susceptibility to male breast cancer in the first instance [66]. Additionally, Nadezhda et al. (2011) found SNPs in FGFR2 and TP53 are cooperatively associated with breast cancer in Caucasian patients, particularly in the postmenopausal period [67]. Thus, SNPs of FGFR2 drive themselves to associate with breast cancer and may contribute to cancer initiation and development. Although our understanding of these SNPs in intron 2 of FGFR2 in breast cancer remains limited, these SNPs may serve as important clinically markers for breast cancer prediction or diagnosis.
Point mutations in FGFR2
Many point mutations in FGFR2 have been reported to cause multiple types of craniosynostoses (Table 2), including Apert syndrome (AS), Crouzon syndrome (CS), Beare-Stevenson syndrome (BS) and Pfeiffer syndrome (PS) etc. Meanwhile, much more mutations were found in various cancers, including those found in craniosynostoses (Figure 1). The cancer types include endometrial carcinomas (12%) [68], lung cancer (3%), skin cancer (rarely), gastric cancer (6%) [69], uterine cancer (rarely), ovarian cancer [70] and breast cancer (4%) [27, 71] (Table 3). Like the point mutations that cause skeletal abnormalities, these point mutations also occur throughout FGFR2, including the ligand-binding and transmembrane domains, and TK domain as well [72].
Nadine et al. (2013) mutated R203C and K660N in FGFR2 and expressed them in HEK293 cells, the result demonstrated both of mutations increased tyrosine kinase activity although K660N mutant was stronger than R203C mutation. Byron et al. (2013) used dovitinib (pan-FGFR inhibitor) to screen for drug-resistant mutations and study the underlying mechanism of drug-resistant [73]. After treating the BaF3 cell line over expressing FGFR2 with high concentration of dovitinib, they identified 14 dovitinib-resistant mutations, including the N550K, which is also observed in breast cancer. Their data demonstrated that the potential mechanism underlying drug resistance is increased receptor tyrosine kinase activity [73].
Mutations in FGFR2 identified in diverse human syndrome
| Human skeletal Disease | Mutation | Ref. |
|---|---|---|
| Apert syndrome (AS) | M186T, P252S/W/F/L, P253L/R, S267P | [103-105] |
| Beare-Stevenson syndrome (BS) | Y375C, S372C | [106] |
| Crouzon syndrome (CS) | A315T/S, A344G/P, C278F, F276V, G338R, K526E, N549D/K/H, Q289P, S267P, S347C, S354C, Y105C, Y281C, Y340C/H | [9, 104, 107-109] |
| Pfeiffer syndrome (PS) | A172F, A314D, C278F, C342F/R/S, K641R, N549D/K, S267P, T341P, W290C, Y340C/H | [9, 104, 110, 111] |
Mutations in FGFR2 identified in diverse human cancers
| Human Cancer Disease | Mutations | Ref. |
|---|---|---|
| Adenoid cystic carcinoma | K642R, Y376C | [109, 112, 113] |
| Bladder cancer | M186T | [114] |
| Breast cancer | R203C, N550K, S588C, K660M | [68, 109, 112, 115, 116] |
| Cervical cancer | A97T, S252L, P256S, K406E, M585V, Y589D, K660M | [68, 117] |
| Colorectal cancer | R203H, R210Q, D334N, Q361R, L552I, P583L, R665W, E778K. | [118-120] |
| Endometrial cancer | D101Y, G227E, S252W, P253R, F276V, K310R, S373C, Y376C, C383R, A390T, M393R, V396D, L398M, I548D/V, N550H/K, K660E/M/N, C383R | [68, 109, 112, 121] |
| Gallbladder cancer | N550K, S252W | [118] |
| Gastric cancer | S267P, Q212K, G463E | [120, 122, 123] |
| Head and neck squamous cell cancer | N550D | [124] |
| Lung cancer | E116K, P253L, I381V, C383R, K421I, D480N, H545Q, G584V, I591M, Q621K, R626T, D138N, N211I, D247Y, D283N, W290C, G302W, S320C, E471Q, M538I, G584W, D603E, K660N/E, L773F, T787K | [102, 125-131] |
| Melanoma | S24F, V77M, H213Y, E219K, G227E, V248D, R251Q, G271E, G305R, T371R, E476K, D531N, E575K, E637K, M641I, I643V, A649T, S689F, G702S, P709S, R760Q, L771V | [132] |
| Oral cancer | V393A, G272V, P253R | [115, 130] |
| Spermatocytic seminoma | S252F/W, P253R/S, S267P, F276V, C278F, Y281C, Q289P, G336R, Y338C/H, C340F/R/S, S352C, K527E, N550K, K642R, K660E | [133] |
Most of these point mutations of FGFR2 result in a gain of function of the protein. The mutations in the extracellular domain enhance the ligand-binding capability and change the ligand specificity [74, 75]. Some mutations in FGFR2 are also known to cause FGFR2 ligand independent dimerization of FGFR2, leading to the activation of its kinase domain [2, 19]. We have had previously introduced the point mutation Ser252Trp (S252W) in mouse FGFR2 and established a mouse model for Apert syndrome (AS) [76]. The mutant mice exhibited severe craniosynostosis characterized by premature fusion of coronal suture, and shortened cranial base. In vitro analysis revealed that the FGFR2-S252W mutation increases apoptosis of osteogenic cells in mutant coronal suture, reduces the space between osteogenic fronts of flat bones, and causes the physical contact of these bones, leading to premature fusion of the coronal suture [76]. Further analysis demonstrated that the S252W mutation activates FGFR2-MAPK-ERK signaling which in turn, triggers bax mediated apoptosis [77].
On the other hand, reports also revealed that R251Q mutation in FGFR2 diminishes its ligand binding affinity [10]. V248D, G227E and G271E mutations impair FGFR2 dimerization, while A648T, D530N and I642V mutations reduce kinase activity [10]. Thus different point mutations of FGFR2 could have different impacts on the functions of FGFR2 and its downstream signaling due to the different nature of the mutations; thus the actual impact of these mutations need to be carefully studied individually.
Strategies for inhibition of FGFR2 signaling as the therapeutic target
Based on the nature of mutations in FGFR2, many approaches can be employed to target FGFR2 signaling for diseases, i.e. targeting FGFR2, the receptors themselves, and downstream signaling molecules. However, as the family of FGFRs share high structural similarity [17], many compounds not only inhibit FGFR2, but other FGFRs as well. Consequently, it is difficult to develop chemical compounds that are highly specific for FGFR2 only. Recently, four inhibitors (ADZ4547, BGJ398, Dovitinib and Lucitanib) have entered phase II trials, and two of them, ADZ4547 and BGJ398, have shown strong specificity to FGFR2 [78-80].
It was shown that FGFR2 signaling mediates resistance to small molecule inhibitors through switching of the HER2 signaling pathway [81]. Studying a cell line, UACC812, which was derived from a lapatinib-resistant breast cancer, Azuma et al. (2011) found these cells contain marked amplification of the FGFR2 gene, which serves as a vital factor for the survival of the resistance breast cancer cell as they become independent of the HER2 pathway [81]. This study suggests that the development of inhibitors to target FGFR2 might become a new therapy strategy to treat HER2-amplified breast cancer patients.
To decrease the side effects of targeting FGFR2, therapeutic antibodies may have much benefit, as they introduced to reduce the potential toxicity of pan-FGFR inhibition for the treatment cancer cells. So far, antibodies targeting FGFR3 have been shown to inhibit bladder cancer and the proliferation of myeloma cells [82, 83], suggesting developing clonal antibodies is a promising field. Otherwise, PDZ173074 and pazopanib showed sensitivity of the FGFR fusion-positive cell line SW780 [84]. It has been shown that breast cancer patients with FGFR2 fusion may benefit from targeted FGFR2 kinase inhibition [85, 86]. Recently, Sommer et al. (2016) developed a novel FGFR2 antibody-drug conjugates (FGFR2-ADC) [87]. In this case, auristatin (a microtubule-disrupting cytotoxic drug) is conjugated to a FGFR2 monoclonal antibody that binds to both FGFR2-IIIb and FGFR2-IIIc isoforms through a non-cleavable linker. Their data demonstrated that the FGFR2-ADC can inhibit growth of FGFR2-positive cell line (MFM-223) and its derived xenograft tumor models with more than 100-fold higher efficiency than FGFR2-negative cell lines [87].
Conclusion and future perspectives
Advances in our understanding of FGFR2 signaling over the past decade have been dramatic. Basic studies into the structure, genetics, cellular biology, and biochemistry of FGFR2 have yielded mounting information regarding how FGFR2 acts through its downstream signaling pathways. The molecular mechanisms underlying activation of FGFR2 is now well understood, but it is also a major challenge to gain more knowledge for future development. Importantly, SNPs in FGFR2 play a vital role in increasing the risk for breast cancer through their interactions with transcription factors to promote tumor formation. Of note, among the 29 SNPs in FGFR2 reviewed here, some of them were associated with luminal A, luminal B (ER/PR+, HER2+), or HER2+/ER- disease, but none were associated with basal-like disease [48, 88]. The underlying mechanisms regarding their association with these particular subtypes of breast cancer is another key challenge for future studies. Furthermore, many point mutations have been found to be associated with various cancers. Although some signaling pathways, such as FGFR2-MAPK-ERK signaling, have been discovered, the mechanisms for each specific mutation for promoting specific types of cancer formation remain elusive. Finally, because large amount of cancers, including breast cancers bear amplification, SNP or point mutations in FGFR2, future efforts should be directed towards the discover of specific inhibitors for FGFR2, and clinical trials of these inhibitors in order to develop effective therapies for FGFR2 associated diseases, including breast cancer.
Acknowledgements
We thank members of the Deng laboratory for critical discussions. This work is not only supported by the Chair Professor Grant (CPG2017-00026-FHS), Startup Research Grant (SRG2015-00045-FHS), MYRG2016-00132-FHS and MYRG2016-00139 of University of Macau and FDCT grants (065/2015/A2 and 094/2015/A3) to Chu-Xia Deng, but also supported by University of Macau, SAR. China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Touat M, Ileana E, Postel-Vinay S, Andre F, Soria J-C. Targeting FGFR Signaling in Cancer. Clinical Cancer Research. 2015;21:2684-94
2. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nature Reviews Cancer. 2010;10:116-29
3. Zhao Y, Adjei AA. Targeting angiogenesis in cancer therapy: moving beyond vascular endothelial growth factor. Oncologist. 2015;20:660-73
4. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139-49
5. Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P. et al. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171-82
6. Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179-85
7. Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM. et al. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res. 2010;70:7630-9
8. Byron SA, Gartside MG, Wellens CL, Mallon MA, Keenan JB, Powell MA. et al. Inhibition of activated fibroblast growth factor receptor 2 in endometrial cancer cells induces cell death despite PTEN abrogation. Cancer Res. 2008;68:6902-7
9. Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446-65
10. Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL. et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Mol Cancer Res. 2009;7:41-54
11. Grose R, Fantl V, Werner S, Chioni AM, Jarosz M, Rudling R. et al. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007;26:1268-78
12. Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci U S A. 2007;104:2973-8
13. Ornitz David M, Marie Pierre J. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463-86
14. Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199-213
15. Tomlinson DC, Knowles MA. Altered Splicing of FGFR1 Is Associated with High Tumor Grade and Stage and Leads to Increased Sensitivity to FGF1 in Bladder Cancer. Am J Pathol. 2010;177:2379-86
16. Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731-42
17. Gong S-G. Isoforms of receptors of fibroblast growth factors. Journal of Cellular Physiology. 2014;229:1887-95
18. Ong SH, Guy GR, Hadari YR, Laks S, Gotoh N, Schlessinger J. et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979-89
19. Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN. et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063-74
20. Ricol D, Cappellen D, El Marjou A, Gil-Diez-de-Medina S, Girault JM, Yoshida T. et al. Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene. 1999;18:7234-43
21. Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Di Bacco A. et al. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340-8
22. Akbarinejad V, Tajik P, Movahedin M, Youssefi R. The role of fibroblast growth factor receptor 2 (FGFR2) in differentiation of bovine spermatogonial stem cells (SCC). Vet Res Forum. 2016;7:149-53
23. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-20
24. Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res. 2010;70:3391-401
25. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90
26. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
27. Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R. et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013-23
28. Sun C, Olopade OI, Di Rienzo A. rs2981582 is associated with FGFR2 expression in normal breast. Cancer Genet Cytogen. 2010;197:193-4
29. Katoh M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol Sci. 2016;37:1081-96
30. Guffanti F, Chila R, Bello E, Zucchetti M, Zangarini M, Ceriani L. et al. In Vitro and In Vivo Activity of Lucitanib in FGFR1/2 Amplified or Mutated Cancer Models. Neoplasia (New York, NY). 2017;19:35-42
31. Sun S, Jiang Y, Zhang G, Song H, Zhang X, Zhang Y. et al. Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2012;105:773-9
32. Lee HJ, Seo AN, Park SY, Kim JY, Park JY, Yu JH. et al. Low Prognostic Implication of Fibroblast Growth Factor Family Activation in Triple-negative Breast Cancer Subsets. Annals of Surgical Oncology. 2014;21:1561-8
33. Huang YL, Chou WC, Hsiung CN, Hu LY, Chu HW, Shen CY. FGFR2 regulates Mre11 expression and double-strand break repair via the MEK-ERK-POU1F1 pathway in breast tumorigenesis. Hum Mol Genet. 2015;24:3506-17
34. Wei W, Liu W, Serra S, Asa SL, Ezzat S. The breast cancer susceptibility FGFR2 provides an alternate mode of HER2 activation. Oncogene. 2015
35. Piasecka D, Kitowska K, Czaplinska D, Mieczkowski K, Mieszkowska M, Turczyk L. et al. Fibroblast growth factor signalling induces loss of progesterone receptor in breast cancer cells. Oncotarget. 2016;7:86011-25
36. Wei WZ, Liu W, Cassol CA, Zheng WY, Asa SL, Ezzat S. The Breast Cancer Susceptibility Gene Product Fibroblast Growth Factor Receptor 2 Serves as a Scaffold for Regulation of NF-kappa B Signaling. Molecular and Cellular Biology. 2012;32:4662-73
37. Kim S, Dubrovska A, Salamone RJ, Walker JR, Grandinetti KB, Bonamy GM. et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PLoS One. 2013;8:e51671
38. Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087-U7
39. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870-4
40. Masuda T, Xu X, Dimitriadis EK, Lahusen T, Deng CX. "DNA Binding Region" of BRCA1 Affects Genetic Stability through modulating the Intra-S-Phase Checkpoint. Int J Biol Sci. 2016;12:133-43
41. Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R. et al. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle. 2015;14:437-48
42. Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T. et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11-20
43. Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416-26
44. Deng CX. Tumor formation in Brca1 conditional mutant mice. Environ Mol Mutagen. 2002;39:171-7
45. Brodie SG, Deng CX. BRCA1-associated tumorigenesis: what have we learned from knockout mice? Trends Genet. 2001;17:S18-22
46. Bane AL, Pinnaduwage D, Colby S, Reedijk M, Egan SE, Bull SB. et al. Expression profiling of familial breast cancers demonstrates higher expression of FGFR2 in BRCA2-associated tumors (vol 117, pg 183, 2009). Breast Cancer Res Tr. 2011;126:269 -
47. Tozlu S, Girault I, Vacher S, Vendrell J, Andrieu C, Spyratos F. et al. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocrine-Related Cancer. 2006;13:1109-20
48. O'Brien KM, Cole SR, Engel LS, Bensen JT, Poole C, Herring AH. et al. Breast cancer subtypes and previously established genetic risk factors: a bayesian approach. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:84-97
49. Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H. et al. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011;13:R110
50. Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL. et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742-54
51. Chen Y, Shi C, Guo Q. TNRC9 rs12443621 and FGFR2 rs2981582 polymorphisms and breast cancer risk. World J Surg Oncol. 2016;14:50
52. Butt S, Harlid S, Borgquist S, Ivarsson M, Landberg G, Dillner J. et al. Genetic predisposition, parity, age at first childbirth and risk for breast cancer. BMC Res Notes. 2012;5:414
53. Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA. et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. Plos Genet. 2008;4:e1000054
54. Liang H, Yang X, Chen L, Li H, Zhu A, Sun M. et al. Heterogeneity of Breast Cancer Associations with Common Genetic Variants in FGFR2 according to the Intrinsic Subtypes in Southern Han Chinese Women. BioMed research international. 2015;2015:626948
55. Han MR, Deming-Halverson S, Cai Q, Wen W, Shrubsole MJ, Shu XO. et al. Evaluating 17 breast cancer susceptibility loci in the Nashville breast health study. Breast cancer (Tokyo, Japan). 2015;22:544-51
56. Andersen SW, Trentham-Dietz A, Figueroa JD, Titus LJ, Cai Q, Long J. et al. Breast cancer susceptibility associated with rs1219648 (fibroblast growth factor receptor 2) and postmenopausal hormone therapy use in a population-based United States study. Menopause (New York, NY). 2013;20:354-8
57. Jara L, Gonzalez-Hormazabal P, Cerceno K, Di Capua GA, Reyes JM, Blanco R. et al. Genetic variants in FGFR2 and MAP3K1 are associated with the risk of familial and early-onset breast cancer in a South-American population. Breast Cancer Res Treat. 2013;137:559-69
58. Shi M, O'Brien KM, Sandler DP, Taylor JA, Zaykin DV, Weinberg CR. Previous GWAS hits in relation to young-onset breast cancer. Breast Cancer Res Treat. 2017;161:333-44
59. Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C. et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108
60. Cox DG, Curtit E, Romieu G, Fumoleau P, Rios M, Bonnefoi H. et al. GWAS in the SIGNAL/PHARE clinical cohort restricts the association between the FGFR2 locus and estrogen receptor status to HER2-negative breast cancer patients. Oncotarget. 2016;7:77358-64
61. Ruiz-Narvaez EA, Haddad SA, Lunetta KL, Yao S, Bensen JT, Sucheston-Campbell LE. et al. Gene-based analysis of the fibroblast growth factor receptor signaling pathway in relation to breast cancer in African American women: the AMBER consortium. Breast Cancer Res Treat. 2016;155:355-63
62. Slattery ML, John EM, Stern MC, Herrick J, Lundgreen A, Giuliano AR. et al. Associations with growth factor genes (FGF1, FGF2, PDGFB, FGFR2, NRG2, EGF, ERBB2) with breast cancer risk and survival: the Breast Cancer Health Disparities Study. Breast Cancer Res Treat. 2013;140:587-601
63. Zhu X, Asa SL, Ezzat S. Histone-acetylated control of fibroblast growth factor receptor 2 intron 2 polymorphisms and isoform splicing in breast cancer. Mol Endocrinol. 2009;23:1397-405
64. Murillo-Zamora E, Moreno-Macias H, Ziv E, Romieu I, Lazcano-Ponce E, Angeles-Llerenas A. et al. Association Between rs2981582 Polymorphism in the FGFR2 Gene and the Risk of Breast Cancer in Mexican Women. Archives of medical research. 2013;44:459-66
65. Xi J, Su Y, Beeghly Fadiel A, Lin Y, Su FX, Jia WH. et al. Association of physical activity and polymorphisms in FGFR2 and DNA methylation related genes with breast cancer risk. Cancer epidemiology. 2014;38:708-14
66. Orr N, Cooke R, Jones M, Fletcher O, Dudbridge F, Chilcott-Burns S. et al. Genetic variants at chromosomes 2q35, 5p12, 6q25.1, 10q26.13, and 16q12.1 influence the risk of breast cancer in men. Plos Genet. 2011;7:e1002290
67. Cherdyntseva NV, Denisov EV, Litviakov NV, Maksimov VN, Malinovskaya EA, Babyshkina NN. et al. Crosstalk between the FGFR2 and TP53 genes in breast cancer: data from an association study and epistatic interaction analysis. DNA and cell biology. 2012;31:306-16
68. Dutt A, Salvesen HB, Chent TH, Ramos AH, Onofrio RC, Hatton C. et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. P Natl Acad Sci USA. 2008;105:8713-7
69. Matsumoto K, Arao T, Hamaguchi T, Shimada Y, Kato K, Oda I. et al. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer. 2012;106:727-32
70. Byron SA, Gartside MG, Wellens CL, Goodfellow PJ, Birrer MJ, Campbell IG. et al. FGFR2 mutations are rare across histologic subtypes of ovarian cancer. Gynecol Oncol. 2010;117:125-9
71. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res. 2016;22:259-67
72. Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C. et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590-2
73. Byron SA, Chen H, Wortmann A, Loch D, Gartside MG, Dehkhoda F. et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia (New York, NY). 2013;15:975-88
74. Zhang XQ, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family - The complete mammalian FGF family. Journal of Biological Chemistry. 2006;281:15694-700
75. Yeh BK, Igarashi M, Eliseenkova AV, Plotnikov AN, Sher I, Ron D. et al. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. P Natl Acad Sci USA. 2003;100:2266-71
76. Chen L, Li D, Li C, Engel A, Deng CX. A Ser252Trp [corrected] substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone. 2003;33:169-78
77. Shukla V, Coumoul X, Wang RH, Kim HS, Deng CX. RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nat Genet. 2007;39:1145-50
78. Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR Signaling in Cancer. Clinical Cancer Research. 2015;21:2684-94
79. Soria JC, DeBraud F, Bahleda R, Adamo B, Andre F, Dienstmann R. et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors (vol 25, pg 2244, 2014). Annals of Oncology. 2015;26:445 -
80. Soria JC, DeBraud F, Bahleda R, Adamo B, Andre F, Dientsmann R. et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Annals of Oncology. 2014;25:2244-51
81. Azuma K, Tsurutani J, Sakai K, Kaneda H, Fujisaka Y, Takeda M. et al. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochemical and biophysical research communications. 2011;407:219-24
82. Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11:6280-90
83. Qing J, Du X, Chen Y, Chan P, Li H, Wu P. et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216-29
84. Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X. et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636-47
85. Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K. et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun. 2015;6:6087
86. Kim SY, Ahn T, Bang H, Ham JS, Kim J, Kim ST. et al. Acquired resistance to LY2874455 in FGFR2-amplified gastric cancer through an emergence of novel FGFR2-ACSL5 fusion. Oncotarget. 2017;8:15014-22
87. Sommer A, Kopitz C, Schatz CA, Nising CF, Mahlert C, Lerchen HG. et al. Preclinical Efficacy of the Auristatin-Based Antibody-Drug Conjugate BAY 1187982 for the Treatment of FGFR2-Positive Solid Tumors. Cancer Res. 2016;76:6331-9
88. O'Brien KM, Cole SR, Poole C, Bensen JT, Herring AH, Engel LS. et al. Replication of breast cancer susceptibility loci in whites and African Americans using a Bayesian approach. Am J Epidemiol. 2014;179:382-94
89. Zhou LP, Yao F, Luan H, Wang YL, Dong XH, Zhou WW. et al. Three novel functional polymorphisms in the promoter of FGFR2 gene and breast cancer risk: a HuGE review and meta-analysis. Breast Cancer Res Tr. 2012;136:885-97
90. Campbell TM, Castro MA, de Santiago I, Fletcher MN, Halim S, Prathalingam R. et al. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis. 2016;37:741-50
91. Hsieh YC, Tu SH, Su CT, Cho EC, Wu CH, Hsieh MC. et al. A polygenic risk score for breast cancer risk in a Taiwanese population. Breast Cancer Res Treat. 2017
92. Xu WH, Shu XO, Long J, Lu W, Cai Q, Zheng Y. et al. Relation of FGFR2 genetic polymorphisms to the association between oral contraceptive use and the risk of breast cancer in Chinese women. Am J Epidemiol. 2011;173:923-31
93. Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K. et al. Incidence of Breast Cancer and Its Subtypes in Relation to Individual and Multiple Low-Penetrance Genetic Susceptibility Loci. Jama-J Am Med Assoc. 2010;304:426-34
94. Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K. et al. Gene-Environment Interactions in 7610 Women With Breast Cancer: Prospective Evidence From the Million Women Study. Obstet Gynecol Surv. 2010;65:704-6
95. Xia P, Li B, Geng T, Deng Z, Dang C, Chang D. et al. FGFR2 gene polymorphisms are associated with breast cancer risk in the Han Chinese population. Am J Cancer Res. 2015;5:1854-61
96. Cui F, Wu DG, Wang WJ, He XT, Wang MH. Variants of FGFR2 and their associations with breast cancer risk: a HUGE systematic review and meta-analysis. Breast Cancer Res Tr. 2016;155:313-35
97. Mazhar A, Jamil F, Bashir Q, Ahmad MS, Masood M, Tanvir I. et al. Genetic variants in FGFR2 and TNRC9 genes are associated with breast cancer risk in Pakistani women. Mol Med Rep. 2016;14:3443-51
98. Nagrani R, Mhatre S, Rajaraman P, Chatterjee N, Akbari MR, Boffetta P. et al. Association of Genome-Wide Association Study (GWAS) Identified SNPs and Risk of Breast Cancer in an Indian Population. Sci Rep. 2017;7:40963
99. Palomba G, Loi A, Porcu E, Cossu A, Zara I, Budroni M. et al. Genome-wide association study of susceptibility loci for breast cancer in Sardinian population. BMC Cancer. 2015;15:383
100. Kim HC, Lee JY, Sung H, Choi JY, Park SK, Lee KM. et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res. 2012;14:R56
101. Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ. et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417-23
102. Chen F, Lu M, Xue Y, Zhou J, Hu F, Chen X. et al. Genetic variants of fibroblast growth factor receptor 2 (FGFR2) are associated with breast cancer risk in Chinese women of the Han nationality. Immunogenetics. 2012;64:71-6
103. Tanimoto Y, Yokozeki M, Hiura K, Matsumoto K, Nakanishi H, Matsumoto T. et al. A soluble form of fibroblast growth factor receptor 2 (FGFR2) with S252W mutation acts as an efficient inhibitor for the enhanced osteoblastic differentiation caused by FGFR2 activation in Apert syndrome. J Biol Chem. 2004;279:45926-34
104. Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. J Cell Physiol. 2012;227:3731-43
105. Morita J, Nakamura M, Kobayashi Y, Deng CX, Funato N, Moriyama K. Soluble form of FGFR2 with S252W partially prevents craniosynostosis of the apert mouse model. Dev Dyn. 2014;243:560-7
106. Slavotinek A, Crawford H, Golabi M, Tao C, Perry H, Oberoi S. et al. Novel FGFR2 deletion in a patient with Beare-Stevenson-like syndrome. Am J Med Genet A. 2009;149A:1814-7
107. Tang M, Huang XJ. Comment on "S267P Mutation in FGFR2: First Report in a Patient With Crouzon Syndrome". J Craniofac Surg. 2015;26:e677
108. Ke R, Yang X, Ge M, Cai T, Lei J, Mu X. S267P mutation in FGFR2: first report in a patient with Crouzon syndrome. J Craniofac Surg. 2015;26:592-4
109. Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H. et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158-62
110. Jay S, Wiberg A, Swan M, Lester T, Williams LJ, Taylor IB. et al. The fibroblast growth factor receptor 2 p.Ala172Phe mutation in Pfeiffer syndrome-history repeating itself. Am J Med Genet A. 2013;161A:1158-63
111. Kelleher FC, O'Sullivan H, Smyth E, McDermott R, Viterbo A. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis. 2013;34:2198-205
112. Byron SA, Gartside M, Powell MA, Wellens CL, Gao F, Mutch DG. et al. FGFR2 point mutations in 466 endometrioid endometrial tumors: relationship with MSI, KRAS, PIK3CA, CTNNB1 mutations and clinicopathological features. PLoS One. 2012;7:e30801
113. Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS. et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;123:2965-8
114. Ruhe JE, Streit S, Hart S, Wong CH, Specht K, Knyazev P. et al. Genetic alterations in the tyrosine kinase transcriptome of human cancer cell lines. Cancer Res. 2007;67:11368-76
115. Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G. et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153-8
116. Reintjes N, Li Y, Becker A, Rohmann E, Schmutzler R, Wollnik B. Activating somatic FGFR2 mutations in breast cancer. PLoS One. 2013;8:e60264
117. Xiang L, Li J, Jiang W, Shen X, Yang W, Wu X. et al. Comprehensive analysis of targetable oncogenic mutations in chinese cervical cancers. Oncotarget. 2015;6:4968-75
118. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70
119. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-7
120. Hansen RM, Goriely A, Wall SA, Roberts IS, Wilkie AO. Fibroblast growth factor receptor 2, gain-of-function mutations, and tumourigenesis: investigating a potential link. The Journal of pathology. 2005;207:27-31
121. Luo F, Xie Y, Xu W, Huang J, Zhou S, Wang Z. et al. Deformed Skull Morphology Is Caused by the Combined Effects of the Maldevelopment of Calvarias, Cranial Base and Brain in FGFR2-P253R Mice Mimicking Human Apert Syndrome. Int J Biol Sci. 2017;13:32-45
122. Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Research. 2001;61:3541-3
123. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061-8
124. Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T. et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632-41
125. Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C. et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713-7
126. Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G. et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153-8
127. Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G. et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591-5
128. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-75
129. Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519-25
130. Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM. et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195-205
131. Tchaicha JH, Akbay EA, Altabef A, Mikse OR, Kikuchi E, Rhee K. et al. Kinase domain activation of FGFR2 yields high-grade lung adenocarcinoma sensitive to a Pan-FGFR inhibitor in a mouse model of NSCLC. Cancer Res. 2014;74:4676-84
132. Gartside MG, Chen HB, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL. et al. Loss-of-Function Fibroblast Growth Factor Receptor-2 Mutations in Melanoma. Mol Cancer Res. 2009;7:41-54
133. Goriely A, Hansen RM, Taylor IB, Olesen IA, Jacobsen GK, McGowan SJ. et al. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat Genet. 2009;41:1247-52
Author contact
![]() Corresponding author: Chu-Xia Deng, Faculty of Health Sciences, University of Macau, Macau SAR, China. cxdengmo
Corresponding author: Chu-Xia Deng, Faculty of Health Sciences, University of Macau, Macau SAR, China. cxdengmo

 Global reach, higher impact
Global reach, higher impact