Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2026; 22(4):2012-2026. doi:10.7150/ijbs.126296 This issue Cite
Review
O-glycosylation in Cancer: Emerging Paradigms and Prospects for Precision Oncology
1. Department of Laboratory Medicine, Jiayu Hospital, Zhongnan Hospital of Wuhan University (People's Hospital of Jiayu County), Xianning 437200, Hubei, China.
2. Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan 430071, Hubei, China.
3. Department of Laboratory Medicine, Huanggang Central Hospital, Huanggang 438000, China.
4. Department of Radiology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, China.
5. Hubei Provincial Clinical Research Center for Molecular Diagnostics, Wuhan 430071, Hubei, China.
#Equally contribution and co-first authors.
Received 2025-10-5; Accepted 2026-1-2; Published 2026-1-22
Abstract
O-glycosylation is a key post-translational modification that profoundly shapes tumor biology by regulating cell proliferation, metastasis, and immune evasion. Aberrant O-glycosylation features truncated glycans such as Tn and sialyl-Tn antigens together with dysregulated glycosyltransferases and promotes oncogenesis in diverse malignancies. This review summarizes recent progress in elucidating the role of O-glycosylation in cancer with emphasis on its effects on cell-surface glycoproteins, intracellular signaling pathways, and emerging RNA modifications. Integration of multi-omics data and machine learning has transformed tumor classification and prognosis prediction through distinct glycosylation signatures and now supports personalized treatment strategies. Newly discovered O-glycosylation of RNA reveals additional regulatory layers and broadens the field of glycosylation research. Targeted interventions including glycosyltransferase inhibitors, gene editing, and combination with immunotherapy demonstrate promising therapeutic potential. Advanced high-throughput tools especially mass spectrometry and enzymatic release methods accelerate biomarker discovery and target validation. Collectively, this review underscores the multifaceted impact of O-glycosylation on cancer progression and treatment response while highlighting the urgent need for continued interdisciplinary collaboration to translate these findings into precision oncology and better patient outcomes.
Keywords: o-glycosylation, protein modification, gene modification, tumor microenvironment, biomarkers
1. Introduction
O-glycosylation represents a major post-translational modification of proteins and regulates diverse biological processes including cell signaling, adhesion, and immune responses. In cancer, aberrant O-glycosylation drives malignant transformation and profoundly affects tumor progression, metastasis, and immune evasion[1]. Dysregulated O-glycans alter protein stability, subcellular localization, and intermolecular interactions, thereby reshaping the functional proteome of cancer cells[1]. The inherent complexity of O-glycosylation, encompassing site-specific alterations and the orchestration by various glycosyltransferases[2], demands deeper mechanistic insight into its oncogenic functions.
In neoplastic cells, disrupted O-glycosylation triggers profound phenotypic reprogramming, promoting uncontrolled proliferation, epithelial-mesenchymal transition (EMT), invasiveness, and suppression of antitumor immunity [1, 3]. These O-glycans activate key oncogenic pathways such as Wnt/β-catenin and phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT), thereby enhancing metastatic potential and therapeutic resistance[4-6].
Recent advances in glycoproteomics, driven by high-resolution mass spectrometry, chemoenzymatic enrichment strategies, and multi-omics integration, have transformed our ability to interrogate site-specific O-glycosylation landscapes with unprecedented depth. These technologies have unveiled cancer-specific glyco-signatures and identified key enzymes, such as polypeptide N-acetylgalactosaminyltransferase 1 (GALNT1), as critical drivers of malignant glyco-phenotypes, highlighting the therapeutic promise of targeting glycosylation machinery to suppress tumor growth and dissemination[7].
O-glycosylation interacts dynamically with other post-translational modifications, especially phosphorylation. These modifications often compete for the same or adjacent serine (Ser)/threonine (Thr) residues and generate complex regulatory networks that control protein function and cell fate in cancer[8]. This review integrates the latest findings on cancer-associated protein O-glycosylation, focusing on molecular mechanisms, functional impact, immune modulation, and clinical translation. By combining protein-centric and genomic views, we aim to establish a solid framework to accelerate glycosylation-based precision oncology strategies.
2. Biosynthesis and Structural Types of O-glycosylation
2.1 Basic Concepts and Classification of O-glycosylation
O-glycosylation represents a pivotal post-translational modification (PTM) characterized by the enzymatic attachment of carbohydrate moieties to the hydroxyl groups of Ser or Thr residues in proteins, facilitated by specialized glycosyltransferases[9]. The inherent complexity of O-glycosylation stems from its lack of consensus sequence motifs and the labile nature of its glycan cores, which hinders site identification. This modification is integral to numerous biological processes, including cell signaling, protein stability, and intercellular interactions[10, 11]. O-glycosylation is broadly divided into mucin-type (O-GalNAc) and O-GlcNAc subtypes. Mucin-type O-GalNAc glycosylation begins with the addition of N-acetylgalactosamine (GalNAc) to Ser/Thr residues and predominates in secreted and membrane-bound mucins that form protective barriers in epithelial tissues[12]. In cancer, mucin-type O-GalNAcylation holds particular clinical importance. The pathway is initiated by the polypeptide N-acetylgalactosaminyltransferase (GALNT) family, which transfer GalNAc to Ser/Thr, forming the Tn antigen. Subsequent extension to core 1 (T antigen) is catalyzed by Core 1 beta1,3-galactosyltransferase (C1GALT1) in complex with its essential chaperone Cosmc. Loss-of-function mutations or epigenetic silencing of C1GALT1 or Cosmc, or aberrant GALNT expression cause accumulation of truncated structures such as Tn and sialyl-Tn (STn) antigens. These oncofetal markers strongly associate with enhanced proliferation, immune evasion, metastasis, and poor prognosis in most epithelial cancers [3, 13, 14] (Figure 1).
2.2 Key Enzymes and Their Regulation
Mucin-type O-glycan biosynthesis depends on a coordinated enzyme network in which GALNT isoforms and C1GALT1 play central roles[13]. C1GALT1, or core 1 β1,3-galactosyltransferase, is indispensable for synthesizing the core 1 motif by transferring galactose to GalNAc, thereby enabling glycan chain elongation. Its functionality relies on the chaperone Cosmc for proper folding, and perturbations in C1GALT1 expression have been linked to aberrant glycosylation in cancers, driving tumor progression[13]. The GALNT family comprises 20 isoforms with distinct yet partially overlapping substrate specificities, peptide preferences, and subcellular localizations. This diversity enables precise spatiotemporal control of glycan initiation[14].
GALNT expression is dynamically regulated by growth factors, oncogenic pathways, hypoxia, and metabolic stress, resulting in profound remodeling of the glycoproteome. Regulation occurs at transcriptional, post-transcriptional, and post-translational levels. Phosphorylation and auto-O-GlcNAcylation directly modulate enzyme activity and stability[15]. Dysregulated signaling often amplifies specific glycosyltransferases, yielding glycosylation profiles that support tumor proliferation, immune escape, and metastasis[16, 17]. For example, GALNT2 overexpression enhances O-glycosylation of growth factor receptors and sustains proliferation within hostile microenvironments [18].
Enzyme competition and cooperation among glycosyltransferases further refine substrate access and final glycan architecture. Alterations in glycosyltransferase levels can generate lectin-recognizable glycans that mediate cell adhesion, signaling, and immunity[13, 19]. Truncated or sialylated O-glycans on tumor surfaces mask antigens, impair immune attack, and facilitate metastasis. Consequently, these enzymes emerge as valuable diagnostic markers and therapeutic targets[20].
2.3 Diversity of O-glycosylation and Tumor-Specific Glycans
Truncated O-glycans, particularly the Tn antigen and STn, represent the most clinically relevant aberrations in cancer[21]. These abbreviated structures are dramatically upregulated in cancers, where they correlate with aggressive phenotypes and unfavorable prognoses [22]. Functionally, Tn/STn promote oncogenic signaling, inhibit apoptosis (including TRAIL-induced cell death), and drive epithelial-mesenchymal transition [23, 24]. Their aberrant patterns also position them as biomarkers for early detection and immunotherapy targets[25].
Glycan heterogeneity varies widely across tumor cell types and influences signaling, adhesion, and immune interactions[26]. Prostate cancers, for example, have unique glycosyltransferase profiles that yield distinct truncated O-glycans compared to normal prostate tissue[27]. Such differences modulate lectin recognition and immune cell trafficking, thereby shaping pro-tumorigenic microenvironments [28]. Comprehensive mapping of O-glycan diversity is therefore essential for precision diagnostics and therapy (Table 1).
In summary, the cancer-associated shift toward truncated and aberrant O-glycans serves as both a hallmark and active driver of malignancy. Deciphering the responsible enzymes and their regulation provides critical opportunities for early detection, accurate prognostication, and development of glycosylation-directed therapeutics.
3. O-glycosylated Proteins in Tumors
3.1 Abnormal O-glycosylation of Cell Surface Glycoproteins and Tumor Progression
Dysregulated O-glycosylation of cell-surface and secreted glycoproteins is a major driver of tumor progression, primarily through altered cell adhesion, migration, and invasion. Cancer cells frequently display truncated O-glycans that profoundly change glycoprotein function. For instance, aberrant O-glycosylation of LGALS3BP enhances tumor cell binding to extracellular matrix and promotes invasion and metastasis[34-36]. Similarly, altered O-glycosylation of cysteine rich with epidermal growth factor (EGF)-like domains 2 (CRELD2) disrupts normal protein interactions and sustains oncogenic signaling[37]. These glycoproteins often bear truncated O-glycans, such as the Tn antigen and its sialylated derivatives, which are prevalent in diverse malignancies and enable immune evasion while exacerbating tumor aggressiveness[43]. Engagement with cognate lectins or receptors further activates proliferative and survival pathways[38].
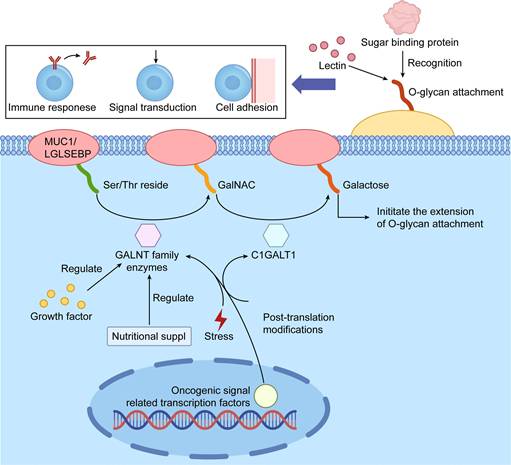
Schematic Illustration of Protein O-glycosylation Initiation and Biological Functions. The figure outlines the key initiation steps of protein O-glycosylation and its critical roles in cellular processes. Specific GALNT family enzymes (N-acetylgalactosaminyltransferases) catalyze the attachment of N-acetylgalactosamine (GalNAc) to Ser or Thr residues, initiating O-glycan chain elongation. This process is regulated by factors such as nutrient availability and cellular stress and can be modulated by other post-translational modifications. The resulting O-glycan structures are recognized by lectins and other glycan-binding proteins, regulating essential processes like cell adhesion, signal transduction, and immune responses, which are pivotal in tumor development and progression.
Functions of Aberrant O-glycosylation
| O-glycosylation | Associated Molecules/Antigens | Role in Tumor Biology | Related Cancers | References |
|---|---|---|---|---|
| Tn antigen expression | MUC1, CD44 | Promotes EMT, migration, and metastasis | Breast cancer | [29, 30] |
| STn antigen expression | TAG-72, MUC16 | Enhances immune evasion, suppresses T-cell infiltration | Colorectal cancer, Ovarian cancer | [31, 32] |
| O-GlcNAc modification | C-Myc | Regulates the tricarboxylic acid cycle | Colorectal cancer | [33] |
| Aberrant O-glycosylation of LGALS3BP | Galectin-3 binding protein (LGALS3BP) | Enhances adhesion to extracellular matrix, promotes invasion and metastasis | Multiple solid tumors | [34-36] |
| Aberrant O-glycosylation of CRELD2 | CRELD2 | Alters protein interactions, activates oncogenic signaling pathways | Various cancers | [37] |
| Aberrant mucin glycosylation | MUC1, MUC4 | Promotes proliferation, metastasis, and immune evasion; MUC1 linked to apoptosis resistance | Breast, Pancreatic, Ovarian, Colorectal cancers | [38] |
| RNA O-glycosylation | O-glycosylated miR-103a-3p, miR-424-5p | Modulates miRNA stability and function; regulates PI3K-Akt signaling; impacts tumor cell growth and metastatic potential | Liver cancer, Lung cancer | [39, 40] |
| Truncated O-glycans | Tn antigen, STn antigen | Facilitate immune evasion, contribute to aggressive phenotype | Multiple cancers | [24, 41] |
| Over-sialylated O-glycans | MUC1, MUC16 | Enhance tumor immune evasion, promote metastasis | Lung cancer | [42] |
Mucins are the most extensively O-glycosylated glycoproteins and play central roles in malignancy. Overexpression and hypoglycosylation of MUC1 and MUC4 create neoepitopes that abolish apical-basolateral polarity, boost proliferation, and confer apoptosis resistance[30, 44, 45]. Aberrant mucin glycoforms also bind siglecs and galectins on immune and stromal cells, thereby remodeling the tumor microenvironment to favor cancer progression[46-48]. Together, these alterations convert cell-surface glycoproteins into active drivers of malignancy, making their modifying enzymes attractive therapeutic targets (Figure 2).
3.2 Intracellular O-GlcNAcylation and Oncogenic Signaling
O-GlcNAcylation dynamically modifies nuclear and cytoplasmic proteins and exerts major effects on oncogenic signaling. O-GlcNAcylation of enolase-1 (ENO1) alters its activity and stability, thereby reprogramming glycolytic flux in cancer cells[3]. In ovarian cancer, this modification enhances cell migration and invasion by shifting phosphorylation patterns[49]. Conversely, O-GlcNAcylation of the p53 protein influences its transcriptional efficacy and stability, potentially stabilizing the protein to favor tumorigenesis in specific contexts[49, 50]. This bifunctional nature underscores O-GlcNAcylation's dual role in tumorigenesis.
O-GlcNAcylation also functions as a nutrient sensor that integrates glucose, amino acid, and lipid metabolism via the hexosamine biosynthetic pathway (HBP)[51]. Elevated UDP-GlcNAc levels in cancer amplify O-GlcNAcylation of metabolic enzymes and transcription factors [52, 53]. This adaptation not only bolsters glycolysis but also aids microenvironmental acclimation, facilitating immune circumvention. O-GlcNAcylation stabilizes PD-L1 through three mechanisms:(1) enhanced transcription via modified STAT3 and NF-κB[54-56], (2) direct modification that blocks ubiquitination and proteasomal degradation[56, 57], and (3) reduced endocytic recycling and lysosomal turnover, resulting in sustained surface expression[58, 59]. These effects make the HBP-OGT axis a central amplifier of PD-1/PD-L1 signaling.
In the immune compartment, aberrant O-GlcNAcylation in tumor-associated macrophages drives M2 polarization and immunosuppressive metabolism[60, 61]. Inhibition of OGT or the HBP therefore destabilizes oncogenic proteins and PD-L1 in tumor cells while simultaneously restoring antitumor immunity, providing strong rationale for combination with PD-1/PD-L1 blockade[62, 63].
3.3 O-glycosylation and the Tumor Immune Microenvironment
Aberrant O-glycosylation reshapes tumor-immune crosstalk within the tumor immune microenvironment (TIME)(Figure 3). Aberrant modifications on tumor cells influence engagements with macrophages and cytotoxic T lymphocytes (CTLs)[6, 64]. Tn antigen expression, for instance, polarizes macrophages toward an M2 phenotype and suppresses antitumor immunity[65]. These glycan patterns also impair CTL recognition and killing, contributing to primary or acquired resistance to checkpoint inhibitors[66, 67].
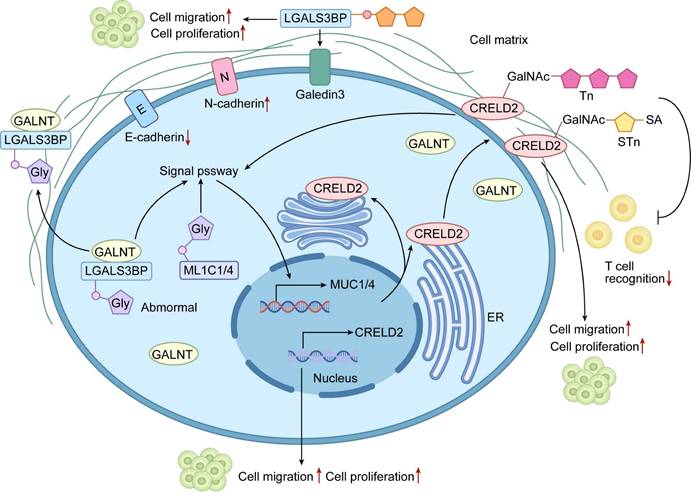
Mechanisms of Aberrant O-glycosylation of Cell Surface Glycoproteins Driving Malignant Phenotypes in Tumors. The diagram elucidates how aberrant O-glycosylation promotes tumor progression by modifying specific cell surface glycoproteins (e.g., LGALS3BP, CRELD2, and MUC1/4). In tumor cells, dysregulated expression or activity of GALNT family enzymes leads to aberrant O-glycosylation. These modifications alter glycoprotein function: LGALS3BP glycosylation enhances tumor cell adhesion to the extracellular matrix, promoting migration and invasion; CRELD2 glycosylation modulates protein interactions, activating oncogenic signaling; and MUC1/4 mucin glycosylation disrupts signaling and adhesion properties. These changes collectively drive core malignant phenotypes, including enhanced proliferation, migration, invasion, and EMT (depicted as E-cadherin to N-cadherin transition), ultimately contributing to tumor progression and metastasis.
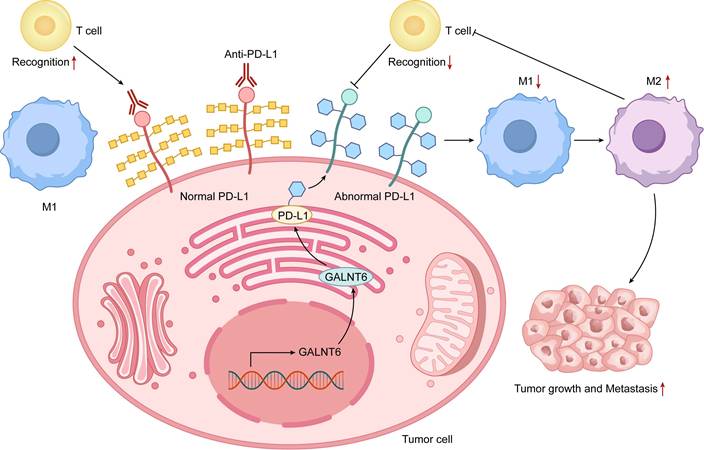
Interactive Diagram of O-glycosylation Regulating the Tumor Immune Microenvironment. The diagram illustrates the dynamic interplay between tumor cells with aberrant O-glycosylation and key immune microenvironment components (T cells and macrophages). The left panel depicts normal immune recognition: unmodified PD-L1 is effectively recognized by T cells, and anti-PD-L1 antibodies function successfully, with macrophages maintaining an antitumor M1 phenotype. The right panel shows an immunosuppressive microenvironment driven by aberrant O-glycosylation (via enzymes like GALNT6): glycosylated PD-L1 on tumor cells impairs T-cell recognition and may reduce immune checkpoint inhibitor efficacy, while glycosylation signals promote macrophage polarization to a protumor M2 phenotype. These mechanisms synergistically drive immune evasion, tumor growth, and metastasis, highlighting O-glycosylation's central role in tumor-immune interactions.
Combining glycosylation inhibitors with immune checkpoint blockade emerges as a powerful synergistic strategy[68]. Inhibition of GALNT6 in pancreatic cancer increases cytotoxic T-cell and macrophage infiltration and enhances immune attack[66]. Such approaches simultaneously restore immune surveillance and directly target tumor cells[69]. As understanding of glycosylation-mediated immune modulation deepens, targeting aberrant O-glycans is gaining traction as a new pillar of cancer immunotherapy (Table 2).
4. O-glycosylation-Related Genes and Their Expression and Function in Tumors
4.1 Expression Profiles and Regulation of O-glycosylation-Related Genes
Recent studies reveal widespread dysregulation of O-glycosylation genes across tumor types and highlight their central role in cancer biology[75-77]. In hepatocellular carcinoma (HCC), transcriptomic analyses show striking changes in GALNT family members compared with adjacent normal liver. These expression patterns faithfully mirror molecular HCC subtypes and strongly predict clinical outcome[77]. Similar upregulation of C1GALT1 and specific GALNTs has been documented in colorectal and breast carcinomas, where high expression correlates with aggressive behavior, immune exclusion, and poor prognosis[78-80]. These findings underscore the need for detailed mechanistic studies of glyco-gene regulation (Figure 4).
Transcriptional control is tightly coupled to the tumor microenvironment. Hypoxia-inducible factor-1α (HIF-1α), activated in hypoxic niches, directly transactivates several GALNTs, driving the synthesis of truncated O-glycans that facilitates metastasis[81]. Extensive crosstalk with Wnt/β-catenin, MYC, and other oncogenic pathways further amplifies this response[82]. Dysregulated O-glycosylation genes and their upstream regulators thus represent promising therapeutic targets to improve treatment efficacy (Table 3).
O-glycosylation in Immune Microenvironment
| Target/Process | Regulation | Immune Consequence | References |
|---|---|---|---|
| PD-L1 glycosylation | Activation (↑) | Suppresses T-cell activity → Immune evasion | [66, 70] |
| GALNT6 activity | Upregulation | Reduces CTL and macrophage infiltration; promotes immunosuppressive TIME | [66] |
| Tn antigen expression | Aberrant O-glycosylation | Facilitates immune evasion; promotes tolerogenic microenvironment | [71, 72] |
| Macrophage polarization | M2 phenotype (↑) | Pro-tumor inflammation, angiogenesis | [65] |
| CTL recognition of tumor cells | Altered by glycosylation | Decreased cytotoxic function | [6] |
| Sialic acid-Siglec binding | Enhancement (↑) | Inhibits NK cell cytotoxicity | [28, 73] |
| O-glycosylation of tumor cell surface | Aberrant | Modulates interactions with immune cells; promotes immune evasion | [74] |
Functional Mechanisms of GALNT Family Genes in Tumor Progression. The mechanistic diagram delineates the central role of GALNT family genes in tumor progression. Under tumor microenvironment stimuli (e.g., hypoxia), transcription factor HIF-1α is activated, upregulating GALNT1 and other GALNT genes. These genes encode GALNTs that catalyze O-glycosylation of downstream substrates (e.g.,EGFR). This post-translational modification enhances receptor stability, sustaining activation of intracellular signaling pathways that promote tumor cell proliferation, survival, and metastasis. The diagram clearly maps the signaling axis from microenvironmental cues to gene expression, protein modification, and malignant phenotype manifestation.
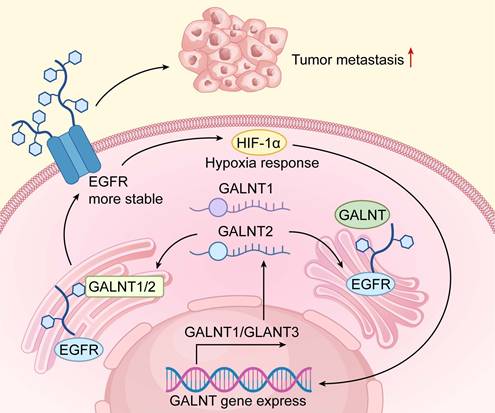
Regulatory Function of GALNT Family
| Gene/miRNA | Regulatory Mechanism | Downstream Effect | Cancer Type & Prognostic Link | References |
|---|---|---|---|---|
| GALNT1 | CD44 glycoproteins modified with abundant Tn antigens | Activating the Wnt/β-catenin signaling pathway | Gastric cancer; higher GALNT1 expression had poorer OS and disease-free survival | [7] |
| GALNT2 | Modulates EGFR expression | Promotes cancer cell proliferation and survival; correlates with HCC molecular subtypes | HCC; linked to tumor progression and prognosis | [83] |
| GALNT6 | Alters O-glycosylation pathways | Lenvatinib resistance; impacts immune cell infiltration and therapy sensitivity | HCC; high expression correlates with drug resistance and treatment stratification | [84] |
| C1GALT1 | Upregulated in tumors | Facilitates immune evasion and tumor growth; affects immune microenvironment | Prostate and breast cancers; associated with aggressive behavior and Tn-positive tumors | [78] |
| COSMC (C1GALT1C1) | Mutation disrupts O-glycosylation | Aberrant Tn antigen expression; promotes tumor growth and immune evasion | Breast cancer; linked to poor prognosis | [41] |
| T-synthase (C1GALT1) | Mutation disrupts O-glycosylation | Tn antigen accumulation; altered gene expression promoting metastasis | Breast cancer; associated with aggressive phenotype | [80] |
| OGT (O-GlcNAc transferase) | Modulates O-glycosylation of miR-424-5p | Influences expression of oncogenes and tumor suppressor genes | Liver cancer; affects tumor progression | [39] |
| GALNT8 | Modulates O-glycosylation affecting EGFR signaling | Suppresses metastatic potential | Breast cancer; regulates tumor metastasis | [85] |
| GALNT14 | Initiates O-GalNAcylation | Upregulates oncogenic factors, promotes proliferation and invasion | Lung adenocarcinoma; promotes tumor growth | [20] |
| miR-103a-3p | O-glycosylation captured by TnORNA method | Regulates cancer cell proliferation and metastatic potential via PI3K-Akt pathway | Pancreatic cancer; aberrant expression linked to tumor growth and metastasis | [86] |
| miR-122-5p | O-glycosylation captured by TnORNA method | Modulates cancer cell proliferation and survival via PI3K-Akt pathway | Pancreatic cancer; aberrant expression promotes tumor progression | [86] |
| miR-424-5p | O-glycosylation modulated through interaction with O-GlcNAc transferase (OGT) | Influences expression of oncogenes and tumor suppressor genes | Liver cancer; affects cancer progression | [39] |
4.2 O-glycosylation Genes in Molecular Subtyping of Tumors
Integration of O-glycosylation gene expression with machine learning has transformed tumor classification, especially in HCC. Consensus clustering using GALNT1, GALNT2, GALNT6, and related genes defines robust molecular subclasses that differ in proliferation rate, immune infiltration, and survival[83, 87, 88]. GALNT1 and GALNT2 directly modulate EGFR O-glycosylation and downstream signaling strength, providing a clear mechanistic link to oncogenic driver activity[83].
Single-cell analyses further resolve intratumoral heterogeneity and predict treatment response. High GALNT6 expression, for example, strongly correlates with lenvatinib resistance in HCC, while distinct glyco-subtypes show variable sensitivity to immune checkpoint blockade[84]. These results position O-glycosylation gene panels as clinically valuable tools for precision oncology and patient stratification.
4.3 Mutations in O-Glycosylation Genes and Tumorigenesis
Somatic mutations and epigenetic silencing of core O-glycosylation genes, most notably Core 1 beta 3-Gal-T-Specific Molecular Chaperone (COSMC /C1GALT1C1) and C1GALT1, are recurrent events in cancers and represent a direct genetic mechanism for the exposure of truncated O-glycans[89]. In triple-negative breast cancer (TNBC), COSMC mutations drive Tn antigen overexpression, which not only serves as a diagnostic marker but also actively reshapes the immune microenvironment[41]. Tn-positive tumors recruit immunosuppressive macrophages through galectin and siglec interactions while suppressing cytotoxic T-cell activity [72]. These alterations confer enhanced metastatic potential and broad therapeutic resistance. Genetic disruption of O-glycosylation pathways therefore emerges as a fundamental oncogenic driver with major implications for tumor progression and clinical management.
5. O-glycosylation of RNA
5.1 Discovery and Detection of RNA O-Glycosylation
The discovery of cell-surface glycosylated RNAs (glycoRNAs) in mammalian cells has revealed a novel class of biomolecules that intersects glycobiology and RNA biology[90]. Although their physiological functions remain poorly defined, initial evidence points to potential roles in cell-cell communication and immune recognition.
Mechanisms of RNA O-glycosylation in Promoting Tumor Progression through the PI3K-Akt Pathway. Schematic illustration depicting the molecular mechanisms by which RNA O-glycosylation drives tumor progression via sustained activation of the PI3K-Akt pathway. In tumor cells, the long non-coding RNA XIST modulates miR-424-5p expression, which in turn regulates the activity of key glycosylation enzymes, including O-GlcNAc transferase (OGT). Concurrently, O-glycosylation of specific miRNAs (e.g., miR-103a-3p and miR-122-5p, markedly enhancing their stability and prolonging their functional lifespan. These glycosylated miRNAs promote persistent PI3K/AKT pathway activation by suppressing tumor suppressor genes and/or negative regulators. The resulting hyperactivation of the PI3K-Akt signaling cascade ultimately orchestrates multiple pro-tumorigenic phenotypes, including enhanced angiogenesis, increased cell survival, and accelerated tumor cell proliferation, collectively driving tumor progression and metastatic dissemination.
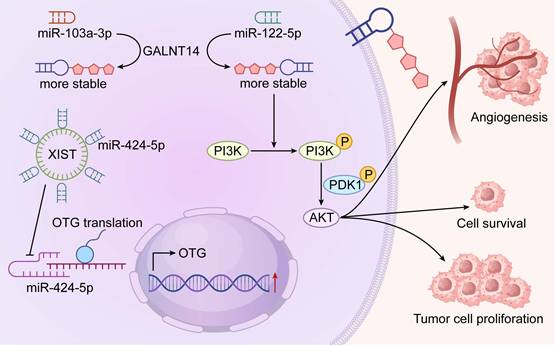
Technological advances have driven early progress in this emerging field. The Tn-containing O-glycosylated RNAs (TnORNA) method stands out as a pioneering chemoenzymatic approach for capturing and enriching O-glycosylated RNA[86, 90]. Using this approach, several miRNAs including miR-103a-3p and miR-122-5p have been identified as O-glycosylated in pancreatic cancer models, with suggested effects on PI3K-Akt signaling and proliferation[86]. Complementary computational tools such as PONglyRNA now predict glycosylation sites on RNA with high confidence and support experimental design[86, 91]. Despite these advances, the stoichiometry, tissue distribution, and functional impact of RNA O-glycosylation are still inadequately characterized. Independent replication of initial findings remains essential.
5.2 O-glycosylated miRNAs in Tumors
Limited but intriguing data indicate that certain miRNAs undergo direct O-glycosylation, which may influence their stability, localization, or target engagement. In pancreatic ductal adenocarcinoma, TnORNA studies report O-glycan attachment to miR-103a-3p and miR-122-5p. These modifications correlate with increased PI3K-Akt activation and enhanced proliferation [86]. Similarly, in HCC, dysregulation of the miR-424-5p/OGT axis has been implicated in oncogenic networks, though direct RNA glycosylation in this context requires verification [39].
Current evidence is primarily descriptive. The consequences of O-glycan addition to miRNAs—such as protection from degradation, altered argonaute binding, or extracellular interactions—remain unproven. The enzymes that catalyze RNA O-glycosylation or remove these marks are still unknown. Thus, while O-glycosylated miRNAs represent an exciting concept, the field is in its infancy. Assertions of clear oncogenic functions should await rigorous mechanistic validation.
5.3 RNA O-glycosylation and Tumor Signaling Pathways
RNA O-glycosylation emerges as a new frontier in cancer biology with potential links to key signaling pathways such as PI3K-Akt (Figure 5). Reported glycosylation of miR-103a-3p and miR-122-5p in pancreatic and lung adenocarcinoma correlates with sustained pathway activity and chemoresistance [40, 86]. In lung cancer, GALNT14-driven protein O-glycosylation similarly boosts proliferation, prompting questions about coordinated regulation of protein and RNA glycosylation [20]. By contrast, GALNT8-mediated glycosylation in breast cancer suppresses metastasis by dampening EGFR signaling and highlights context-specific effects[85].
Computational tools now facilitate transcriptome-wide prediction of glycosylation sites and guide targeted investigations[86]. However, causal evidence connecting RNA O-glycosylation to altered signaling remains limited. The field requires prioritized efforts in independent validation, enzyme identification, and precise functional studies to distinguish correlation from true regulatory mechanisms.
6. O-glycosylation as a Target for Tumor Diagnosis and Therapy
6.1 O-glycosylation-Related Biomarkers
Tumor-associated carbohydrate antigens (TACAs) generated by aberrant O-glycosylation, including Tn, STn, and CA19-9, serve as established diagnostic and prognostic markers. Tn antigen is overexpressed in many adenocarcinomas and detectable in tissue biopsies and serum, allowing non-invasive monitoring of tumor burden and therapy response. STn similarly predicts aggressive disease and poor outcome, while CA19-9 remains the standard serum marker for pancreatic cancer [42, 92]. Salivary glycoproteomics has identified distinct sialylation and fucosylation patterns in early lung cancer, supporting non-invasive screening approaches[93, 94]. High-resolution mass spectrometry platforms now enable site-specific and quantitative glycan analysis, driving discovery of more sensitive and specific biomarkers[95, 96] (Table 4).
Clinical translation of O-glycosylation biomarkers faces substantial challenges. Glycosylation patterns exhibit profound heterogeneity at multiple levels. Inter-patient variation arises from genetics, microbiome, and metabolism[100]. Intra-tumor heterogeneity, driven by subclonal diversity and microenvironmental gradients such as hypoxia, creates spatially variable glyco-profiles[28, 101]. This mosaicism causes sampling bias in biopsies and dilutes tumor signals in liquid biopsies. Truncated glycans like Tn also appear in benign inflammation, reducing specificity[102]. Technical hurdles include O-glycan lability, low site occupancy, scarcity of reliable site-specific antibodies, and limited clinical compatibility of glycoproteomics workflows.
6.2 Therapeutic Targeting of O-glycosylation
Pharmacological inhibition of O-glycosylation emerges as a promising adjuvant to immunotherapy. Repurposed itraconazole suppresses glycosylation in head and neck cancer and markedly improves anti-PD-1 response by shifting macrophages to an M1 phenotype, enhancing CD8+ T-cell activity, and lowering immunosuppressive cytokines[6]. These benefits arise from remodeled tumor-surface glycans that disrupt inhibitory immune checkpoints.
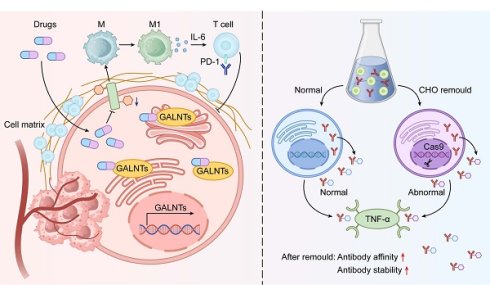
Gene editing and enzyme modulation offer additional avenues. CRISPR/Cas9 knockout of C1GALT1 induces Tn/STn exposure, disrupts oncogenic signaling, and sensitizes colorectal and pancreatic tumors to immune clearance[98]. Selective glycosyltransferase inhibitors alter tumor antigen glycans to boost immune recognition[103] (Figure 6). These strategies simultaneously impair tumor cell fitness and overcome immune evasion. Continued development of O-glycosylation-targeted therapies holds potential for personalized treatment across cancer types (Table 5).
6.3 O-glycosylation Engineering Cell Platforms and Biotherapeutic Optimization
Glycoengineering of O-glycosylation in producer cells has become essential for optimizing biopharmaceuticals. Specialized Chinese hamster ovary (CHO) platforms now allow precise control of O-glycan structures on recombinant proteins[106]. These systems produce variants with defined glycans that influence activity and pharmacokinetics. For example, etanercept analogs bearing truncated (Tn/STn) or extended (sialyl-core 1/3) O-glycans show improved TNF-α binding and therapeutic efficacy[99]. Such findings establish O-glycosylation as a critical quality attribute in biotherapeutic development.
O-glycosylation-Derived Diagnostic Biomarkers
| Biomarker | Sample Type | Clinical Utility | References |
|---|---|---|---|
| Tn/STn antigens | Serum, Tissue | Early screening, prognosis evaluation | [71] |
| Sialylated glycoproteins | Saliva | Non-invasive lung cancer screening | [42] |
| Glycan profiles detected by mass spectrometry | Body fluids (e.g., saliva, serum) | Detailed characterization of glycosylation changes for novel biomarker identification | [97] |
| PD-L1 glycosylation | Tumor tissue | Predictive biomarker for immunotherapy (anti-PD-1/PD-L1 efficacy) | [66] |
| M2 macrophage-associated O-glycosylation signatures | Tumor microenvironment | Indicator of immunosuppressive tumor state | [65] |
| Sialic acid-Siglec binding patterns | Tumor tissue, immune cells | Biomarker of NK cell suppression and immune escape | [28] |
| C1GALT1 downregulation-associated O-glycosylation profile | Colorectal cancer tissue | Associated with tumor aggressiveness; potential prognostic biomarker | [98] |
| Etanercept variants with distinct O-glycosylation patterns (sialylTn, Tn, sialylCore 3, sialylCore 1) | Engineered therapeutic protein (CHO platform) | Quality attribute biomarker for therapeutic efficacy | [99] |
| O-glycosylation pattern | Tumor tissue | Serves as a predictive biomarker for response to combination therapy with itraconazole and anti-PD-1 | [6] |
Schematic of Therapeutic Strategies Targeting O-glycosylation. The diagram integrates two key therapeutic strategies targeting tumor O-glycosylation. The left panel depicts small molecule inhibitors that suppress GALNT enzyme activity, blocking aberrant O-glycosylation and reversing immunosuppressive tumor microenvironments. This approach synergy with immune checkpoint inhibitors. The right panel showcases glycosylation engineering platforms, using CRISPR/Cas9-modified CHO cells to optimize therapeutic proteins (e.g., antibodies or receptor fusion proteins) by tailoring O-glycosylation, improving binding affinity and stability to targets like TNF-α. These strategies collectively highlight the multidimensional therapeutic potential of targeting O-glycosylation from microenvironment modulation to biotherapeutic optimization.
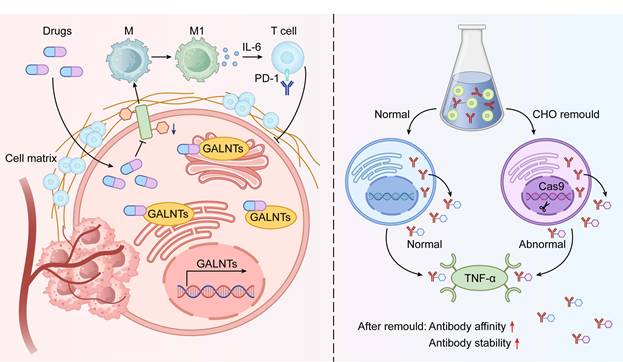
Strategies Targeting O-glycosylation
| Strategy | Representative Approach | Mechanism of Action | References |
|---|---|---|---|
| Therapeutic Targeting of Tumor-Specific Glycopeptides | Development of monoclonal antibodies targeting glycopeptide epitopes | Specific targeting of glycopeptides to enhance tumor cell recognition while minimizing damage to healthy tissue | [74] |
| Glycosyltransferase Inhibitors | Itraconazole | Blocks GALNT activity → Inhibits PD-L1 glycosylation | [6] |
| Glycoengineering | CHO cell platform | Optimizes O-glycan structures → Improves binding affinity | [99] |
| Gene Editing | CRISPR-mediated GALNT14 knockdown | Suppresses RNA O-GalNAc modification → Blocks PI3K pathway | [20] |
| Glycosylation-Targeted Cancer Vaccines | Glycopeptide-based vaccines targeting tumor-associated carbohydrate antigens | Induces immune responses against tumor-specific glycosylation markers, aiding in the recognition and destruction of tumor cells | [104] |
| Glycosylation inhibitors | Disrupt tumor-associated glycocalyx | Disrupts the protective glycocalyx formed by sialylated O-glycans on tumor cells, enhancing immune recognition | [6] |
| RNA Glycosylation | Targeting glycosylated RNA (glycoRNA) | Regulates immune responses and tumor progression | [86, 105] |
Selective O-glycan modulation preserves N-glycosylation and overall protein integrity[13]. This approach enhances performance while enabling systematic study of glycan effects on product quality. Broader application will improve diverse biologics and deliver superior patient outcomes. Robust glycoengineered platforms are therefore vital for advancing biopharmaceutical manufacturing to modern standards.
7. Technological Advances and Future Research Directions
7.1 Development of High-Throughput Analysis Technologies
High-throughput technologies have revolutionized glycosylation research in oncology. Ultrahigh-resolution mass spectrometry, including MALDI-FT-ICR MS, enables simultaneous profiling of N- and O-glycans. These methods provide detailed structural analysis of complex glycans from cancer cell lines and tissues. A semi-automated workflow has profiled O-glycans in colorectal and pancreatic cancer cells, identifying compositions from monosaccharides to branched oligosaccharides[97]. Automated tools like MassyTools improve quantification accuracy using signal-to-noise and mass error criteria across large datasets[97].
Glycopeptide enrichment strategies have further advanced O-glycosylation analysis. The MOTAI method combines enzymatic digestion with solid-phase extraction to boost sensitivity and coverage in tumor samples[37]. This approach has identified upregulated Tn/STn-glycoproteins in colorectal cancer compared with normal tissue, highlighting diagnostic potential. Sequential O-glycoprotease digestion in MOTAI isolates O-GalNAc sites and elucidates aberrant glycoprotein functions [37].
Integration with high-throughput sequencing and bioinformatics correlates glycan profiles with clinical outcomes [107]. Ongoing refinements will deepen insights into glycosylation-cancer interactions and refine diagnostic paradigms[107, 108].
7.2 Multi-omics and Machine Learning Applications in O-glycosylation
Multi-omics integration elucidates O-glycosylation networks in tumors. Weighted gene co-expression network analysis (WGCNA) in HCC identifies key glyco-genes and defines molecular subtypes linked to behavior and prognosis[77]. Subtype CS1 shows genomic heterogeneity and moderate immune infiltration. Subtype CS2 exhibits genomic stability and favorable immune profiles [77]. Combining glycomics with genomics and transcriptomics reveals O-glycosylation effects on the tumor microenvironment, immune phenotypes, and outcomes, aiding biomarker and target discovery [77, 109].
Machine learning enhances O-glycosylation-based classification and prognostication[110]. Evaluation of 59 algorithms has yielded robust HCC prognostic models from glyco-gene expression, defining clinically relevant subtypes[77]. CS2 patients display superior survival and strong immune infiltration, predicting immunotherapy response. CS3 patients face poor prognosis with genomic instability, requiring alternative approaches[77]. These machine-learning signatures support clinical decision-making and patient stratification. Multi-omics and machine learning thus drive personalized oncology for HCC and beyond[77].
7.3 New Opportunities for O-glycosylation Modification in Tumor Immunotherapy
O-glycosylation regulates tumor immunobiology by modulating immune checkpoints and cell function. Aberrant O-glycans on tumor glycoproteins stabilize PD-L1, impair T-cell activation, and promote immune evasion [56]. Targeting these modifications may boost checkpoint inhibitor efficacy. Glycopeptide epitopes from CD44v6 have enabled development of selective monoclonal antibodies with enhanced tumor targeting and safety[74, 111]. Personalized glycopeptide vaccines mimicking TACAs could elicit strong antitumor immunity and overcome resistance[104].
Sialylated O-glycans form a protective glycocalyx that shields tumors from immune attack[112]. Inhibitors or glycan-engineered immune cells that disrupt sialylation may synergize with immunotherapies[113]. Glycosylation profiles also predict immunotherapy response and guide treatment selection [114, 115]. The immunomodulatory roles of O-glycosylation merit intensive study to advance cancer immunotherapy and patient outcomes.
7.4 Future Exploration in RNA Glycosylation
RNA glycosylation represents a new regulatory layer with major implications for cancer. GlycoRNAs on small RNAs modulate immune interactions and tumor progression[90, 116, 117]. Aberrant glycoRNA patterns associate with malignancy and resistance[117, 118]. Coexistence of N- and O-linked modifications on RNAs suggests interplay with protein glycosylation, influencing gene regulation and signaling[86]. Future studies should define glycoRNA functions and cross-talk with protein pathways to uncover therapeutic avenues.
Advanced detection technologies are essential for progress. Current methods lack sensitivity, necessitating tools like solid-phase chemoenzymatic gRNA (SPCgRNA) for selective capture and profiling [105]. Bioinformatics predictors such as PONglyRNA identify RNA glycosylation sites for hypothesis testing[86]. Extension to long non-coding RNAs, circular RNAs, and other classes will clarify roles in disease.
Therapeutic targeting of RNA glycosylation offers novel oncology strategies. Altering glycoRNA pathways may influence RNA stability, cellular localization, and downstream signaling, thereby affecting tumor proliferation, apoptosis, and immune evasion[117, 119]. Multi-omics integration will decode the cancer glyco-code and enable personalized approaches. Sustained RNA glycosylation research promises key insights and innovative treatments.
8. Conclusion
Glycosylation profoundly influences tumor biology by regulating proliferation, metastasis, and immune interactions. Aberrant forms, characterized by truncated glycans such as Tn and STn antigens alongside dysregulated glycosyltransferases, actively drive oncogenesis and disease progression. Integration of multi-omics data with machine learning has transformed tumor subtyping, prognostic modeling, and treatment selection. These tools uncover cancer-specific glycosylation signatures and enable personalized interventions to improve outcomes.
The discovery of RNA O-glycosylation introduces an exciting new regulatory layer. GlycoRNAs hold promise as diagnostic markers, prognostic indicators, and therapeutic targets, warranting intensive study to integrate them into precision oncology. Therapeutically, targeted modulation of aberrant O-glycosylation, particularly in combination with immunotherapy, yields promising preclinical and early clinical results that may expand treatment efficacy.
Significant barriers persist in clinical translation. The inherent complexity of O-glycans—including low stoichiometry, heterogeneity, and technical detection challenges—necessitates continued advances in mass spectrometry, enrichment strategies, and analytical workflows. Large-scale validation studies and interdisciplinary collaboration among glycobiologists, oncologists, immunologists, bioinformaticians, and clinicians remain crucial. Overcoming these obstacles will unlock the full potential of O-glycosylation research to deliver innovative diagnostic tools and transformative therapies in precision oncology.
Abbreviations
EMT: Epithelial-mesenchymal transition; PI3K/AKT: Phosphatidylinositol 3-kinase /protein kinase B; GALNT1: Polypeptide N-acetylgalactosaminyl-transferase 1; PTMs: post-translational modifications; Ser: Serine; Thr: Threonine; O-GalNAc: Mucin-type O-linked N-acetylgalactosamine; GalNAc: N-acetylgalactosamine; GALNT: Polypeptide N-acetylgalactosaminyltransferase; C1GALT1: Core 1 beta 1,3-galactosyltransferase; STn: Sialyl Tn antigen; GalNAc-T: N-acetylgalactosaminyltransferase; LGALS3BP: Galectin-3-Binding Protein; CRELD2: cysteine rich with epidermal growth factor (EGF)-like domains 2; MUC1: Mucin 1; ENO1: Elpha-Enolase; HBP: Hexosamine biosynthetic pathway; PD-L1: Programmed cell death ligand 1; STAT3: Signal transducer and activator of transcription 3; NF-kappaB: Nuclear Factor Kappa-light-chain-enhancer of Activated B Cells; PDL-1: Programmed death ligand 1; PD-1: programmed cell death protein-1; OGT: O-GlcNAc transferase; TIME: Tumor immune microenvironment; CTLs: Tytotoxic T lymphocytes; PDAC: Pancreatic Ductal Adenocarcinoma; HCC: Hepatocellular carcinoma; HIF-1α: Hypoxia-inducible factor 1α; EGFR: Epidermal growth factor receptor; COSMC /C1GALT1C1: Core 1 beta 3-Gal-T Specific Molecular Chaperone; TNBC: Triple negative breast cancer; GlycoRNAs: Glycosylated RNAs; TnORNA: Tn-containing O-glycosylated RNAs; PONglyRNA: Predictor of O-and N-linked glycoRNA; EGFR: Epidermal growth factor receptor; TACAs: Tumor-associated carbohydrate antigens; CHO: Chinese hamster ovary; TNF-α: Tumor necrosis factor-alpha; MALDI-FT-ICR: Matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance; WGCNA: Weighted gene co-expression network analysis; CS1: cancer subtype 1; CD44v6: Cluster of differentiation 44 variant 6; SPCgRNA: Solid-phase chemoenzymatic gRNA.
Acknowledgements
This work was supported by the Hubei Provincial Youth Talents Program for Public Health (WSJKRC2022013) and the Yellow Crane Talent Program of Wuhan City (HHYC2019002).
Author Contributions
Junhao Wei: Visualization, Writing- Original draft preparation. Shengbao Hu: Visualization, Writing- Original draft preparation. Wanfang Chen: Writing- Original draft preparation. Hye Song Paek: Writing- Original draft preparation. Guohong Liu: Conceptualization, Validation, Writing- Original draft preparation, Funding acquisition. Yunbao Pan: Conceptualization, Validation, Supervision, Writing- Original draft preparation, Writing - Review & Editing, Funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nature reviews Cancer. 2015;15:540-55
2. Hashii N, Suzuki J. Site-Specific O-Glycosylation Analysis by Liquid Chromatography-Mass Spectrometry with Electron-Transfer/Higher-Energy Collisional Dissociation. Methods in molecular biology (Clifton, NJ). 2021;2271:169-78
3. Zhu Q, Li J, Sun H, Fan Z, Hu J, Chai S. et al. O-GlcNAcylation of enolase 1 serves as a dual regulator of aerobic glycolysis and immune evasion in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2024;121:e2408354121
4. Liu Z, Liu J, Dong X, Hu X, Jiang Y, Li L. et al. Tn antigen promotes human colorectal cancer metastasis via H-Ras mediated epithelial-mesenchymal transition activation. Journal of cellular and molecular medicine. 2019;23:2083-92
5. Rosa-Fernandes L, Oba-Shinjo SM, Macedo-da-Silva J, Marie SKN, Palmisano G. Aberrant Protein Glycosylation in Brain Cancers, with Emphasis on Glioblastoma. Advances in experimental medicine and biology. 2022;1382:39-70
6. Lin MC, Chuang YT, Wu HY, Hsu CL, Lin NY, Huang MC. et al. Targeting tumor O-glycosylation modulates cancer-immune-cell crosstalk and enhances anti-PD-1 immunotherapy in head and neck cancer. Molecular oncology. 2024;18:350-68
7. Zhang J, Wang H, Wu J, Yuan C, Chen S, Liu S. et al. GALNT1 Enhances Malignant Phenotype of Gastric Cancer via Modulating CD44 Glycosylation to Activate the Wnt/β-catenin Signaling Pathway. International journal of biological sciences. 2022;18:6068-83
8. Şener Uslupehlivan E, Deveci R, Şahar U, İzzetoğlu S. Glycan analysis of Lamin A/C protein at G2/M and S phases of the cell cycle. Cell biochemistry and biophysics. 2022;80:689-98
9. Zhou F, Ma J, Zhu Y, Wang T, Yang Y, Sun Y. et al. The role and potential mechanism of O-Glycosylation in gastrointestinal tumors. Pharmacological research. 2022;184:106420
10. Thompson N, Wakarchuk W. O-glycosylation and its role in therapeutic proteins. Bioscience reports. 2022 42
11. Chatham JC, Zhang J, Wende AR. Role of O-Linked N-Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiological reviews. 2021;101:427-93
12. Carraway KL, Hull SR. O-glycosylation pathway for mucin-type glycoproteins. BioEssays: news and reviews in molecular, cellular and developmental biology. 1989;10:117-21
13. Sun X, Zhan M, Sun X, Liu W, Meng X. C1GALT1 in health and disease. Oncology letters. 2021;22:589
14. Nielsen MI, de Haan N, Kightlinger W, Ye Z, Dabelsteen S, Li M. et al. Global mapping of GalNAc-T isoform-specificities and O-glycosylation site-occupancy in a tissue-forming human cell line. Nature communications. 2022;13:6257
15. Yan S, Yuan K, Yao X, Chen Q, Li J, Sun J. 14-3-3ε augments OGT stability by binding with S20-phosphorylated OGT. The Journal of biological chemistry. 2024;300:107774
16. Le Minh G, Esquea EM, Young RG, Huang J, Reginato MJ. On a sugar high: Role of O-GlcNAcylation in cancer. The Journal of biological chemistry. 2023;299:105344
17. Cai M, Chen Y, Lin Y, Hu Z, Li L, Huang H. et al. SIRT1 Asn346 sugar chain promoting collagen deacetylation protective effect on osteoblasts under stress. Biochemical and biophysical research communications. 2023;682:148-55
18. Dong X, Leng Y, Tian T, Hu Q, Chen S, Liu Y. et al. GALNT2, an O-glycosylating enzyme, is a critical regulator of radioresistance of non-small cell lung cancer: evidence from an integrated multi-omics analysis. Cell biology and toxicology. 2023;39:3159-74
19. Collette AM, Hassan SA, Schmidt SI, Lara AJ, Yang W, Samara NL. An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase. Science advances. 2024;10:eadj8829
20. Tang B, Wang K, Ren Q, Zhou J, Xu Y, Liu L. et al. GALNT14-mediated O-glycosylation drives lung adenocarcinoma progression by reducing endogenous reactive oxygen species generation. Cellular signalling. 2024;124:111477
21. Holst S, Wuhrer M, Rombouts Y. Glycosylation characteristics of colorectal cancer. Advances in cancer research. 2015;126:203-56
22. Rømer TB, Aasted MKM, Dabelsteen S, Groen A, Schnabel J, Tan E. et al. Mapping of truncated O-glycans in cancers of epithelial and non-epithelial origin. British journal of cancer. 2021;125:1239-50
23. Jiang Y, Wen T, Yan R, Kim SR, Stowell SR, Wang W. et al. O-glycans on death receptors in cells modulate their sensitivity to TRAIL-induced apoptosis through affecting on their stability and oligomerization. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2020;34:11786-801
24. Li R, Dong X, Chen S, Tan J, Chen X, Liu J. et al. Tn antigen promotes breast cancer metastasis via impairment of CASC4. Cell biology international. 2023;47:1854-67
25. Takakura D, Ohashi S, Kobayashi N, Tokuhisa M, Ichikawa Y, Kawasaki N. Targeted O-glycoproteomics for the development of diagnostic markers for advanced colorectal cancer. Frontiers in oncology. 2023;13:1104936
26. Madunić K, Zhang T, Mayboroda OA, Holst S, Stavenhagen K, Jin C. et al. Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cellular and molecular life sciences: CMLS. 2021;78:337-50
27. Hodgson K, Orozco-Moreno M, Scott E, Garnham R, Livermore K, Thomas H. et al. The role of GCNT1 mediated O-glycosylation in aggressive prostate cancer. Scientific reports. 2023;13:17031
28. Rodriguez E. Tumor Glycosylation: A Main Player in the Modulation of Immune Responses. European journal of immunology. 2025;55:e202451318
29. Du T, Jia X, Dong X, Ru X, Li L, Wang Y. et al. Cosmc Disruption-Mediated Aberrant O-glycosylation Suppresses Breast Cancer Cell Growth via Impairment of CD44. Cancer management and research. 2020;12:511-22
30. Xi X, Wang J, Qin Y, Huang W, You Y, Zhan J. Glycosylated modification of MUC1 maybe a new target to promote drug sensitivity and efficacy for breast cancer chemotherapy. Cell death & disease. 2022;13:708
31. Reddish MA, Jackson L, Koganty RR, Qiu D, Hong W, Longenecker BM. Specificities of anti-sialyl-Tn and anti-Tn monoclonal antibodies generated using novel clustered synthetic glycopeptide epitopes. Glycoconjugate journal. 1997;14:549-60
32. Akita K, Yoshida S, Ikehara Y, Shirakawa S, Toda M, Inoue M. et al. Different levels of sialyl-Tn antigen expressed on MUC16 in patients with endometriosis and ovarian cancer. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2012;22:531-8
33. Wang H, Sun J, Sun H, Wang Y, Lin B, Wu L. et al. The OGT-c-Myc-PDK2 axis rewires the TCA cycle and promotes colorectal tumor growth. Cell death and differentiation. 2024;31:1157-69
34. Fogeron ML, Müller H, Schade S, Dreher F, Lehmann V, Kühnel A. et al. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nature communications. 2013;4:1531
35. Pece A, Lovato G, Cela I, Mercatelli A, Ferro B, Nikkola J. et al. Glycosylated LGALS3BP is highly secreted by bladder cancer cells and represents a novel urinary disease biomarker. Molecular oncology. 2025
36. Kimura R, Yoshimaru T, Matsushita Y, Matsuo T, Ono M, Park JH. et al. The GALNT6-LGALS3BP axis promotes breast cancer cell growth. International journal of oncology. 2020;56:581-95
37. Yue S, Wang X, Wang L, Li J, Zhou Y, Chen Y. et al. MOTAI: A Novel Method for the Study of O-GalNAcylation and Complex O-Glycosylation in Cancer. Analytical chemistry. 2024;96:11137-45
38. Brockhausen I, Melamed J. Mucins as anti-cancer targets: perspectives of the glycobiologist. Glycoconjugate journal. 2021;38:459-74
39. Ning D, Chen J, Du P, Liu Q, Cheng Q, Li X. et al. The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. Liver international: official journal of the International Association for the Study of the Liver. 2021;41:1933-44
40. Li H, Huhe M, Lou J. MicroRNA-103a-3p Promotes Cell Proliferation and Invasion in Non-Small-Cell Lung Cancer Cells through Akt Pathway by Targeting PTEN. BioMed research international. 2021;2021:7590976
41. Festari MF, Jara E, Costa M, Iriarte A, Freire T. Truncated O-glycosylation in metastatic triple-negative breast cancer reveals a gene expression signature associated with extracellular matrix and proteolysis. Scientific reports. 2024;14:1809
42. Gao Z, Xu M, Yue S, Shan H, Xia J, Jiang J. et al. Abnormal sialylation and fucosylation of saliva glycoproteins: Characteristics of lung cancer-specific biomarkers. Current research in pharmacology and drug discovery. 2022;3:100079
43. Cervoni GE, Cheng JJ, Stackhouse KA, Heimburg-Molinaro J, Cummings RD. O-glycan recognition and function in mice and human cancers. The Biochemical journal. 2020;477:1541-64
44. Li S, Dong X, Gao K, Wang Y, Li M, Deng H. et al. PP2A Promotes the Symmetric Division of MUC1-Dominant Cancer Stem-Like Cells in Small Cell Lung Cancer. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2025;12:e2503545
45. Chen C, Patel A, Demirkhanyan L, Gondi CS. The Role of Mucins in Cancer and Cancer Progression: A Comprehensive Review. Current issues in molecular biology. 2025 47
46. Ito K, Stannard K, Gabutero E, Clark AM, Neo SY, Onturk S. et al. Galectin-1 as a potent target for cancer therapy: role in the tumor microenvironment. Cancer metastasis reviews. 2012;31:763-78
47. Swanson BJ, McDermott KM, Singh PK, Eggers JP, Crocker PR, Hollingsworth MA. MUC1 is a counter-receptor for myelin-associated glycoprotein (Siglec-4a) and their interaction contributes to adhesion in pancreatic cancer perineural invasion. Cancer research. 2007;67:10222-9
48. Chen W, Cheng Q, Li N, Gu K, Zhao H, Na H. The role of glycan-lectin interactions in the tumor microenvironment: immunosuppression regulators of colorectal cancer. American journal of cancer research. 2025;15:1347-83
49. Song M, Suh PG. O-GlcNAcylation regulates lysophosphatidic acid-induced cell migration by regulating ERM family proteins. FEBS open bio. 2022;12:1220-9
50. Jo S, Kim JS, Baek SH, Jang YK. O-GlcNAcylation of DDB1 at Ser-764 by OGT promotes cancer cell stemness in colorectal cancer through increased polyubiquitination of p53. Scientific reports. 2025
51. Chen L, Hu M, Chen L, Peng Y, Zhang C, Wang X. et al. Targeting O-GlcNAcylation in cancer therapeutic resistance: The sugar Saga continues. Cancer letters. 2024;588:216742
52. Le Minh G, Reginato MJ. Role of O-GlcNAcylation on cancer stem cells: Connecting nutrient sensing to cell plasticity. Advances in cancer research. 2023;157:195-228
53. Liu X, Blaženović I, Contreras AJ, Pham TM, Tabuloc CA, Li YH. et al. Hexosamine biosynthetic pathway and O-GlcNAc-processing enzymes regulate daily rhythms in protein O-GlcNAcylation. Nature communications. 2021;12:4173
54. Fu Y, Ning L, Feng J, Yu X, Guan F, Li X. Dynamic regulation of O-GlcNAcylation and phosphorylation on STAT3 under hypoxia-induced EMT. Cellular signalling. 2022;93:110277
55. Dong H, Liu Z, Wen H. Protein O-GlcNAcylation Regulates Innate Immune Cell Function. Frontiers in immunology. 2022;13:805018
56. Zhu Q, Wang H, Chai S, Xu L, Lin B, Yi W. et al. O-GlcNAcylation promotes tumor immune evasion by inhibiting PD-L1 lysosomal degradation. Proceedings of the National Academy of Sciences of the United States of America. 2023;120:e2216796120
57. Li XM, Wu HL, Xia QD, Zhou P, Wang SG, Yu X. et al. Novel insights into the SPOP E3 ubiquitin ligase: From the regulation of molecular mechanisms to tumorigenesis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2022;149:112882
58. Liu B, Wu Y, Xiang A, Yu Y, Song S, Zhang Y. et al. The hexosamine biosynthetic pathway drives tumor immune evasion via translational control of PD-L1 at the elongation level. Cell reports. 2025;44:116249
59. Itano N, Iwamoto S. Dysregulation of hexosamine biosynthetic pathway wiring metabolic signaling circuits in cancer. Biochimica et biophysica acta General subjects. 2023;1867:130250
60. Li Y, Qu S, Jin H, Jia Q, Li M. Role of O-GlcNAcylation in cancer biology. Pathology, research and practice. 2024;253:155001
61. Rodrigues Mantuano N, Stanczak MA, Oliveira IA, Kirchhammer N, Filardy AA, Monaco G. et al. Hyperglycemia Enhances Cancer Immune Evasion by Inducing Alternative Macrophage Polarization through Increased O-GlcNAcylation. Cancer Immunol Res. 2020;8:1262-72
62. Wang J, Xie Y, Liu L, Rong S, Cai H, Zeng H. et al. O-GlcNAc transferase promotes immune evasion and immunotherapy resistance in uterine corpus endometrial cancer by targeting the glucocorticoid receptor. Journal for immunotherapy of cancer. 2025 13
63. Ouyang M, Yu C, Deng X, Zhang Y, Zhang X, Duan F. O-GlcNAcylation and Its Role in Cancer-Associated Inflammation. Frontiers in immunology. 2022;13:861559
64. Shi Q, Shen Q, Liu Y, Shi Y, Huang W, Wang X. et al. Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote cancer metastasis and chemoresistance. Cancer cell. 2022;40:1207-22.e10
65. Rajesh C, Radhakrishnan P. The (Sialyl) Tn antigen: Contributions to immunosuppression in gastrointestinal cancers. Frontiers in oncology. 2022;12:1093496
66. Chong S, Liu Y, Bian Z, Hu D, Guo S, Dong C. et al. GALNT6 dual regulates innate immunity STING signaling and PD-L1 expression to promote immune evasion in pancreatic ductal adenocarcinoma. Cellular signalling. 2025;134:111942
67. Qusairy Z, Rada M. Glycosylation in cancer: mechanisms, diagnostic markers, and therapeutic applications. Molecular and cellular biochemistry. 2025
68. Zhu Q, Chen X, Duan X, Sun J, Yi W. Targeting glycosylation to enhance tumor immunotherapy. Trends in pharmacological sciences. 2025;46:863-76
69. Ashok G, Soundararajan A, Anbarasu A, Ramaiah S. Elucidating the molecular role of MUC5B in progressive lung adenocarcinoma: Prospects for early diagnosis. Journal of molecular recognition: JMR. 2024;37:e3064
70. Zhang W, Zhao E, Li Z, Liu W, Wang J, Hou W. et al. Hexokinase HK3-mediated O-GlcNAcylation of EP300: a key regulator of PD-L1 expression and immune evasion in ccRCC. Cell death & disease. 2024;15:613
71. Mercanoglu B, Karstens KF, Giannou AD, Meiners J, Lücke J, Seeger P. et al. A Comprehensive Analysis of Tn and STn Antigen Expression in Esophageal Adenocarcinoma. Cancers. 2024 16
72. da Costa V, van Vliet SJ, Carasi P, Frigerio S, García PA, Croci DO. et al. The Tn antigen promotes lung tumor growth by fostering immunosuppression and angiogenesis via interaction with Macrophage Galactose-type lectin 2 (MGL2). Cancer letters. 2021;518:72-81
73. Yang ST, Liu CH, Chao WT, Liu HH, Lee WL, Wang PH. The role of sialylation in gynecologic cancers. Taiwanese journal of obstetrics & gynecology. 2023;62:651-4
74. Aasted MKM, Groen AC, Keane JT, Dabelsteen S, Tan E, Schnabel J. et al. Targeting Solid Cancers with a Cancer-Specific Monoclonal Antibody to Surface Expressed Aberrantly O-glycosylated Proteins. Molecular cancer therapeutics. 2023;22:1204-14
75. García-Vallejo JJ, Gringhuis SI, van Dijk W, van Die I. Gene expression analysis of glycosylation-related genes by real-time polymerase chain reaction. Methods in molecular biology (Clifton, NJ). 2006;347:187-209
76. Zhang Z, Tan EP, VandenHull NJ, Peterson KR, Slawson C. O-GlcNAcase Expression is Sensitive to Changes in O-GlcNAc Homeostasis. Frontiers in endocrinology. 2014;5:206
77. Li M, Li H, Liu L, Wei Q, Gao J, Hu B. et al. Integrated multi-omics analysis and machine learning refine molecular subtypes and prognosis in hepatocellular carcinoma through O-linked glycosylation genes. Functional & integrative genomics. 2025;25:162
78. Kalemoglu E, Caner A. Bioinformatic Analysis of C1GALT1 in Cancer: Insights into Prognosis, Metastasis and Therapeutic Potential. Cancer reports (Hoboken, NJ). 2025;8:e70259
79. Lin NY, Lee JJ, Chen ST, Lin JA, Lin CH, Lin HY. et al. Truncation of GalNAc-type O-glycans Suppresses CD44-mediated Osteoclastogenesis and Bone Metastasis in Breast Cancer. Molecular cancer research: MCR. 2023;21:664-74
80. Chou CH, Huang MJ, Chen CH, Shyu MK, Huang J, Hung JS. et al. Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget. 2015;6:6123-35
81. Wolters-Eisfeld G, Mercanoglu B, Strohmaier A, Guengoer C, Izbicki JR, Bockhorn M. Hypoxia induced HIF1a-mediated O-GalNAc glycosylation of cytosolic O-GlcNAcylated proteins to regulate signaling pathways in pancreatic cancer. Journal of Clinical Oncology. 35: e15739-e.
82. Dietinger V, García de Durango CR, Wiechmann S, Boos SL, Michl M, Neumann J. et al. Wnt-driven LARGE2 mediates laminin-adhesive O-glycosylation in human colonic epithelial cells and colorectal cancer. Cell communication and signaling: CCS. 2020;18:102
83. Osman TE, Guo Y, Li S. Exploring the combined roles of GALNT1 and GALNT2 in hepatocellular carcinoma malignancy and EGFR modulation. Discover oncology. 2025;16:337
84. Zhang P, Chen S, Cai J, Song L, Quan B, Wan J. et al. GALNT6 drives lenvatinib resistance in hepatocellular carcinoma through autophagy and cancer-associated fibroblast activation. Cellular oncology (Dordrecht, Netherlands). 2024;47:2439-60
85. Huang T, Meng F, Huang H, Wang L, Wang L, Liu Y. et al. GALNT8 suppresses breast cancer cell metastasis potential by regulating EGFR O-GalNAcylation. Biochemical and biophysical research communications. 2022;601:16-23
86. Li J, Wang L, Deng J, Miao X, Yang X, Zhou Y. et al. O-Glycosylated RNA Identification and Prediction by Solid-Phase Chemoenzymatic TnORNA Method and PONglyRNA Tool. Analytical chemistry. 2025;97:16906-15
87. Dai T, Li J, Liang RB, Yu H, Lu X, Wang G. Identification and Experimental Validation of the Prognostic Significance and Immunological Correlation of Glycosylation-Related Signature and ST6GALNAC4 in Hepatocellular Carcinoma. Journal of hepatocellular carcinoma. 2023;10:531-51
88. Wang Y, Chen H. Protein glycosylation alterations in hepatocellular carcinoma: function and clinical implications. Oncogene. 2023;42:1970-9
89. Thomas D, Sagar S, Caffrey T, Grandgenett PM, Radhakrishnan P. Truncated O-glycans promote epithelial-to-mesenchymal transition and stemness properties of pancreatic cancer cells. Journal of cellular and molecular medicine. 2019;23:6885-96
90. Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG. et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021;184:3109-24.e22
91. Xie Y, Chai P, Till NA, Hemberger H, Lebedenko CG, Porat J. et al. The modified RNA base acp(3)U is an attachment site for N-glycans in glycoRNA. Cell. 2024;187:5228-37.e12
92. Berois N, Pittini A, Osinaga E. Targeting Tumor Glycans for Cancer Therapy: Successes, Limitations, and Perspectives. Cancers. 2022 14
93. Liu S, Wang H, Jiang X, Ji Y, Wang Z, Zhang Y. et al. Integrated N-glycoproteomics Analysis of Human Saliva for Lung Cancer. Journal of proteome research. 2022;21:1589-602
94. Gao Z, Wu Z, Han Y, Zhang X, Hao P, Xu M. et al. Aberrant Fucosylation of Saliva Glycoprotein Defining Lung Adenocarcinomas Malignancy. ACS omega. 2022;7:17894-906
95. Balbisi M, Sugár S, Turiák L. Protein glycosylation in lung cancer from a mass spectrometry perspective. Mass spectrometry reviews. 2024
96. Nagai-Okatani C, Nishigori M, Sato T, Minamino N, Kaji H, Kuno A. Wisteria floribunda agglutinin staining for the quantitative assessment of cardiac fibrogenic activity in a mouse model of dilated cardiomyopathy. Laboratory investigation; a journal of technical methods and pathology. 2019;99:1749-65
97. Kotsias M, Madunić K, Nicolardi S, Kozak RP, Gardner RA, Jansen BC. et al. A semi-automated, high throughput approach for O-glycosylation profiling of in vitro established cancer cell lines by MALDI-FT-ICR MS. Glycoconjugate journal. 2021;38:747-56
98. Tian H, Yu JL, Chu X, Guan Q, Liu J, Liu Y. Unraveling the role of C1GALT1 in abnormal glycosylation and colorectal cancer progression. Frontiers in oncology. 2024;14:1389713
99. Biel TG, Faison T, Matthews AM, Zou G, Ortega-Rodriguez U, Pegues MA. et al. An etanercept O-glycovariant with enhanced potency. Molecular therapy Methods & clinical development. 2022;25:124-35
100. Wandall HH, Nielsen MAI, King-Smith S, de Haan N, Bagdonaite I. Global functions of O-glycosylation: promises and challenges in O-glycobiology. The FEBS journal. 2021;288:7183-212
101. Peixoto A, Ferreira D, Miranda A, Relvas-Santos M, Freitas R, Veth TS. et al. Multilevel plasticity and altered glycosylation drive aggressiveness in hypoxic and glucose-deprived bladder cancer cells. iScience. 2025;28:111758
102. Ju T, Wang Y, Aryal RP, Lehoux SD, Ding X, Kudelka MR. et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 2013;7:618-31
103. Chi JJ, Xie P, Cheng MH, Zhu Y, Cui X, Watson J. et al. MGAT1-Guided complex N-Glycans on CD73 regulate immune evasion in triple-negative breast cancer. Nature communications. 2025;16:3552
104. Pesce E, Sodini A, Palmieri E, Valensin S, Tinti C, Rossi M. et al. GMMA decorated with mucin 1 Tn/STn mimetics elicit specific antibodies response and inhibit tumor growth. NPJ vaccines. 2025;10:71
105. Li J, Yue S, Gao Z, Hu W, Liu Z, Xu G. et al. Novel Approach to Enriching Glycosylated RNAs: Specific Capture of GlycoRNAs via Solid-Phase Chemistry. Analytical chemistry. 2023;95:11969-77
106. Prati EG, Matasci M, Suter TB, Dinter A, Sburlati AR, Bailey JE. Engineering of coordinated up- and down-regulation of two glycosyltransferases of the O-glycosylation pathway in Chinese hamster ovary (CHO) cells. Biotechnology and bioengineering. 2002;79:580-5
107. Petrović T, Trbojević-Akmačić I. Lectin and Liquid Chromatography-Based Methods for Immunoglobulin (G) Glycosylation Analysis. Experientia supplementum (2012). 2021;112:29-72
108. Puranik A, Saldanha M, Chirmule N, Dandekar P, Jain R. Advanced strategies in glycosylation prediction and control during biopharmaceutical development: Avenues toward industry 4.0. Biotechnology progress. 2022;38:e3283
109. Xie Y, Liu X, Yi L, Wang S, Lin Z, Zhao C. et al. Development and application of GlycanDIA workflow for glycomic analysis. Nature communications. 2025;16:7075
110. Hong S, Chattaraj KG, Guo J, Trout BL, Braatz RD. Enhanced O-glycosylation site prediction using explainable machine learning technique with spatial local environment. Bioinformatics (Oxford, England). 2025 41
111. Porcellini S, Asperti C, Corna S, Cicoria E, Valtolina V, Stornaiuolo A. et al. CAR T Cells Redirected to CD44v6 Control Tumor Growth in Lung and Ovary Adenocarcinoma Bearing Mice. Front Immunol. 2020;11:99
112. Habeeb IF, Alao TE, Delgado D, Buffone A Jr. When a negative (charge) is not a positive: sialylation and its role in cancer mechanics and progression. Frontiers in oncology. 2024;14:1487306
113. Lim J, Sari-Ak D, Bagga T. Siglecs as Therapeutic Targets in Cancer. Biology. 2021 10
114. Xu X, Chen Z, Song M, Hou Z, Balmer L, Zhou C. et al. Profiling of IgG N-glycosylation for axial spondyloarthritis and other rheumatic diseases. Arthritis research & therapy. 2025;27:37
115. Zhang W, Chen T, Zhao H, Ren S. Glycosylation in aging and neurodegenerative diseases. Acta biochimica et biophysica Sinica. 2024;56:1208-20
116. Deng J, Miao X, Wang X, Wen SY, Zhou Z, Li S. et al. Galactose oxidase oxidation and glycosidase digestion for glycoRNA analysis. Analytical methods: advancing methods and applications. 2025;17:964-71
117. Kim HS. GlycoRNA: A new player in cellular communication. Oncology research. 2025;33:995-1000
118. Zheng L, Yang Q, Li F, Zhu M, Yang H, Tan T. et al. The Glycosylation of Immune Checkpoints and Their Applications in Oncology. Pharmaceuticals (Basel, Switzerland). 2022 15
119. Graziano VR, Porat J, Ah Kioon MD, Mejdrová I, Matz AJ, Lebedenko CG. et al. RNA N-glycosylation enables immune evasion and homeostatic efferocytosis. Nature. 2025;645:784-92
Author contact
![]() Corresponding authors: Guohong Liu, Department of Radiology, Zhongnan Hospital of Wuhan University, Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan, 430071, China; E-mail: liuguohcom; Yunbao Pan, Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan, 430071, China; E-mail: panyunbaocom.
Corresponding authors: Guohong Liu, Department of Radiology, Zhongnan Hospital of Wuhan University, Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan, 430071, China; E-mail: liuguohcom; Yunbao Pan, Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan, 430071, China; E-mail: panyunbaocom.

 Global reach, higher impact
Global reach, higher impact